Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1
- PMID: 33846315
- PMCID: PMC8041777
- DOI: 10.1038/s41467-021-22320-y
Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1
Abstract
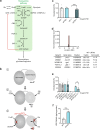
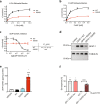
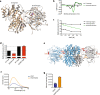
The hexosamine pathway (HP) is a key anabolic pathway whose product uridine 5'-diphospho-N-acetyl-D-glucosamine (UDP-GlcNAc) is an essential precursor for glycosylation processes in mammals. It modulates the ER stress response and HP activation extends lifespan in Caenorhabditis elegans. The highly conserved glutamine fructose-6-phosphate amidotransferase 1 (GFAT-1) is the rate-limiting HP enzyme. GFAT-1 activity is modulated by UDP-GlcNAc feedback inhibition and via phosphorylation by protein kinase A (PKA). Molecular consequences of GFAT-1 phosphorylation, however, remain poorly understood. Here, we identify the GFAT-1 R203H substitution that elevates UDP-GlcNAc levels in C. elegans. In human GFAT-1, the R203H substitution interferes with UDP-GlcNAc inhibition and with PKA-mediated Ser205 phosphorylation. Our data indicate that phosphorylation affects the interactions of the two GFAT-1 domains to control catalytic activity. Notably, Ser205 phosphorylation has two discernible effects: it lowers baseline GFAT-1 activity and abolishes UDP-GlcNAc feedback inhibition. PKA controls the HP by uncoupling the metabolic feedback loop of GFAT-1.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.Nat Commun. 2020 Feb 4;11(1):687. doi: 10.1038/s41467-020-14524-5. Nat Commun. 2020. PMID: 32019926 Free PMC article.
-
Phosphorylation of human glutamine:fructose-6-phosphate amidotransferase by cAMP-dependent protein kinase at serine 205 blocks the enzyme activity.J Biol Chem. 2000 Jul 21;275(29):21981-7. doi: 10.1074/jbc.M001049200. J Biol Chem. 2000. PMID: 10806197
-
Hexosamine pathway and (ER) protein quality control.Curr Opin Cell Biol. 2015 Apr;33:14-8. doi: 10.1016/j.ceb.2014.10.001. Epub 2014 Oct 28. Curr Opin Cell Biol. 2015. PMID: 25463841 Review.
-
Mapping the UDP-N-acetylglucosamine regulatory site of human glucosamine-6P synthase by saturation-transfer difference NMR and site-directed mutagenesis.Biochimie. 2014 Feb;97:39-48. doi: 10.1016/j.biochi.2013.09.011. Epub 2013 Sep 26. Biochimie. 2014. PMID: 24075873
-
Hexosamines, insulin resistance, and the complications of diabetes: current status.Am J Physiol Endocrinol Metab. 2006 Jan;290(1):E1-E8. doi: 10.1152/ajpendo.00329.2005. Am J Physiol Endocrinol Metab. 2006. PMID: 16339923 Free PMC article. Review.
Cited by
-
Hexosamine pathway activation improves memory but does not extend lifespan in mice.Aging Cell. 2022 Oct;21(10):e13711. doi: 10.1111/acel.13711. Epub 2022 Sep 19. Aging Cell. 2022. PMID: 36124412 Free PMC article.
-
The UDPase ENTPD5 regulates ER stress-associated renal injury by mediating protein N-glycosylation.Cell Death Dis. 2023 Feb 27;14(2):166. doi: 10.1038/s41419-023-05685-4. Cell Death Dis. 2023. PMID: 36849424 Free PMC article.
-
Inhibitors of glucosamine-6-phosphate synthase as potential antimicrobials or antidiabetics - synthesis and properties.J Enzyme Inhib Med Chem. 2022 Dec;37(1):1928-1956. doi: 10.1080/14756366.2022.2096018. J Enzyme Inhib Med Chem. 2022. PMID: 35801410 Free PMC article.
-
Roles of GFAT and PFK genes in energy metabolism of brown planthopper, Nilaparvata lugens.Front Physiol. 2023 Jun 21;14:1213654. doi: 10.3389/fphys.2023.1213654. eCollection 2023. Front Physiol. 2023. PMID: 37415905 Free PMC article.
-
Slt2 Is Required to Activate ER-Stress-Protective Mechanisms through TORC1 Inhibition and Hexosamine Pathway Activation.J Fungi (Basel). 2022 Jan 18;8(2):92. doi: 10.3390/jof8020092. J Fungi (Basel). 2022. PMID: 35205847 Free PMC article.
References
-
- Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J. Biol. Chem. 1991;266:4706–4712. doi: 10.1016/S0021-9258(19)67706-9. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

