The white matter is a pro-differentiative niche for glioblastoma
- PMID: 33846316
- PMCID: PMC8042097
- DOI: 10.1038/s41467-021-22225-w
The white matter is a pro-differentiative niche for glioblastoma
Erratum in
-
Author Correction: The white matter is a pro-differentiative niche for glioblastoma.Nat Commun. 2022 Apr 14;13(1):2163. doi: 10.1038/s41467-022-29916-y. Nat Commun. 2022. PMID: 35422055 Free PMC article. No abstract available.
Abstract
Glioblastomas are hierarchically organised tumours driven by glioma stem cells that retain partial differentiation potential. Glioma stem cells are maintained in specialised microenvironments, but whether, or how, they undergo lineage progression outside of these niches remains unclear. Here we identify the white matter as a differentiative niche for glioblastomas with oligodendrocyte lineage competency. Tumour cells in contact with white matter acquire pre-oligodendrocyte fate, resulting in decreased proliferation and invasion. Differentiation is a response to white matter injury, which is caused by tumour infiltration itself in a tumoursuppressive feedback loop. Mechanistically, tumour cell differentiation is driven by selective white matter upregulation of SOX10, a master regulator of normal oligodendrogenesis. SOX10 overexpression or treatment with myelination-promoting agents that upregulate endogenous SOX10, mimic this response, leading to niche-independent pre-oligodendrocyte differentiation and tumour suppression in vivo. Thus, glioblastoma recapitulates an injury response and exploiting this latent programme may offer treatment opportunities for a subset of patients.
Conflict of interest statement
The authors declare no competing interests.
Figures


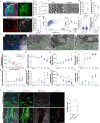
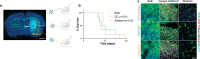

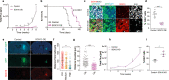

References
Publication types
MeSH terms
Substances
Grants and funding
- G0701018/MRC_/Medical Research Council/United Kingdom
- 28990/CRUK_/Cancer Research UK/United Kingdom
- MR/N004272/1/MRC_/Medical Research Council/United Kingdom
- 17368/CRUK_/Cancer Research UK/United Kingdom
- MC_UP_1102/8/MRC_/Medical Research Council/United Kingdom
- 23459/CRUK_/Cancer Research UK/United Kingdom
- 21992/CRUK_/Cancer Research UK/United Kingdom
- 27590/CRUK_/Cancer Research UK/United Kingdom
- 21203/CRUK_/Cancer Research UK/United Kingdom
- MC_U12266B/MRC_/Medical Research Council/United Kingdom
- G1100578/MRC_/Medical Research Council/United Kingdom
- MC-A658-5TY20/MRC_/Medical Research Council/United Kingdom
- A21992/CRUK_/Cancer Research UK/United Kingdom
- 28592/CRUK_/Cancer Research UK/United Kingdom
- MC_UP_A652_1002/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

