Learning cis-regulatory principles of ADAR-based RNA editing from CRISPR-mediated mutagenesis
- PMID: 33846332
- PMCID: PMC8041805
- DOI: 10.1038/s41467-021-22489-2
Learning cis-regulatory principles of ADAR-based RNA editing from CRISPR-mediated mutagenesis
Abstract
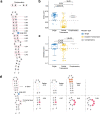
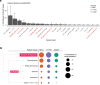
Adenosine-to-inosine (A-to-I) RNA editing catalyzed by ADAR enzymes occurs in double-stranded RNAs. Despite a compelling need towards predictive understanding of natural and engineered editing events, how the RNA sequence and structure determine the editing efficiency and specificity (i.e., cis-regulation) is poorly understood. We apply a CRISPR/Cas9-mediated saturation mutagenesis approach to generate libraries of mutations near three natural editing substrates at their endogenous genomic loci. We use machine learning to integrate diverse RNA sequence and structure features to model editing levels measured by deep sequencing. We confirm known features and identify new features important for RNA editing. Training and testing XGBoost algorithm within the same substrate yield models that explain 68 to 86 percent of substrate-specific variation in editing levels. However, the models do not generalize across substrates, suggesting complex and context-dependent regulation patterns. Our integrative approach can be applied to larger scale experiments towards deciphering the RNA editing code.
Conflict of interest statement
The authors declare the following competing interests: J.B.L. is a co-founder of AIRNA Bio and a consultant for Risen Pharma. Anna Shcherbina receives consulting fees from Myokardia, Inc, is a scientific adviser to Ravel Bio, Inc., and an employee of Insitro, Inc.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

