Developing a non-destructive metabarcoding protocol for detection of pest insects in bulk trap catches
- PMID: 33846382
- PMCID: PMC8041782
- DOI: 10.1038/s41598-021-85855-6
Developing a non-destructive metabarcoding protocol for detection of pest insects in bulk trap catches
Abstract
Metabarcoding has the potential to revolutionise insect surveillance by providing high-throughput and cost-effective species identification of all specimens within mixed trap catches. Nevertheless, incorporation of metabarcoding into insect diagnostic laboratories will first require the development and evaluation of protocols that adhere to the specialised regulatory requirements of invasive species surveillance. In this study, we develop a multi-locus non-destructive metabarcoding protocol that allows sensitive detection of agricultural pests, and subsequent confirmation using traditional diagnostic techniques. We validate this protocol for the detection of tomato potato psyllid (Bactericera cockerelli) and Russian wheat aphid (Diuraphis noxia) within mock communities and field survey traps. We find that metabarcoding can reliably detect target insects within mixed community samples, including specimens that morphological identification did not initially detect, but sensitivity appears inversely related to community size and is impacted by primer biases, target loci, and sample indexing strategy. While our multi-locus approach allowed independent validation of target detection, lack of reference sequences for 18S and 12S restricted its usefulness for estimating diversity in field samples. The non-destructive DNA extraction proved invaluable for resolving inconsistencies between morphological and metabarcoding identification results, and post-extraction specimens were suitable for both morphological re-examination and DNA re-extraction for confirmatory barcoding.
Conflict of interest statement
The authors declare no competing interests.
Figures

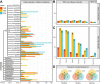
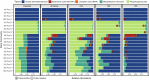


Similar articles
-
Prospects and challenges of implementing DNA metabarcoding for high-throughput insect surveillance.Gigascience. 2019 Aug 1;8(8):giz092. doi: 10.1093/gigascience/giz092. Gigascience. 2019. PMID: 31363753 Free PMC article. Review.
-
Disentangling bias for non-destructive insect metabarcoding.PeerJ. 2022 Feb 23;10:e12981. doi: 10.7717/peerj.12981. eCollection 2022. PeerJ. 2022. PMID: 35228909 Free PMC article.
-
Biodiversity assessments in the 21st century: the potential of insect traps to complement environmental samples for estimating eukaryotic and prokaryotic diversity using high-throughput DNA metabarcoding 1.Genome. 2019 Mar;62(3):147-159. doi: 10.1139/gen-2018-0096. Epub 2019 Jan 23. Genome. 2019. PMID: 30673361
-
Establishing arthropod community composition using metabarcoding: Surprising inconsistencies between soil samples and preservative ethanol and homogenate from Malaise trap catches.Mol Ecol Resour. 2019 Nov;19(6):1516-1530. doi: 10.1111/1755-0998.13071. Epub 2019 Sep 18. Mol Ecol Resour. 2019. PMID: 31379089 Free PMC article.
-
Bioinformatic challenges for DNA metabarcoding of plants and animals.Mol Ecol. 2012 Apr;21(8):1834-47. doi: 10.1111/j.1365-294X.2012.05550.x. Mol Ecol. 2012. PMID: 22486822 Review.
Cited by
-
Development of a DNA Metabarcoding Method for the Identification of Insects in Food.Foods. 2023 Mar 3;12(5):1086. doi: 10.3390/foods12051086. Foods. 2023. PMID: 36900603 Free PMC article.
-
FAVIS: Fast and versatile protocol for non-destructive metabarcoding of bulk insect samples.PLoS One. 2023 Jul 19;18(7):e0286272. doi: 10.1371/journal.pone.0286272. eCollection 2023. PLoS One. 2023. PMID: 37467453 Free PMC article.
-
Validating a multi-locus metabarcoding approach for characterizing mixed-pollen samples.Plant Methods. 2023 Nov 4;19(1):120. doi: 10.1186/s13007-023-01097-9. Plant Methods. 2023. PMID: 37925401 Free PMC article.
-
Repeated subsamples during DNA extraction reveal increased diversity estimates in DNA metabarcoding of Malaise traps.Ecol Evol. 2022 Nov 27;12(11):e9502. doi: 10.1002/ece3.9502. eCollection 2022 Nov. Ecol Evol. 2022. PMID: 36447594 Free PMC article.
-
Specific gut bacterial responses to natural diets of tropical birds.Sci Rep. 2022 Jan 13;12(1):713. doi: 10.1038/s41598-022-04808-9. Sci Rep. 2022. PMID: 35027664 Free PMC article.
References
-
- Deiner K, Bik HM, Mächler E, Seymour M, Lacoursière-Roussel A, Altermatt F, Creer S, Bista I, Lodge DM, de Vere N, Pfrender ME, Bernatchez L. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017;26(21):5872–5895. doi: 10.1111/mec.14350. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

