Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2
- PMID: 33846513
- PMCID: PMC8041869
- DOI: 10.1038/s42003-021-02030-3
Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2
Abstract
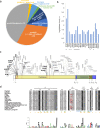
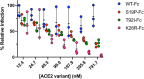
COVID-19 is a respiratory illness caused by a novel coronavirus called SARS-CoV-2. The viral spike (S) protein engages the human angiotensin-converting enzyme 2 (ACE2) receptor to invade host cells with ~10-15-fold higher affinity compared to SARS-CoV S-protein, making it highly infectious. Here, we assessed if ACE2 polymorphisms can alter host susceptibility to SARS-CoV-2 by affecting this interaction. We analyzed over 290,000 samples representing >400 population groups from public genomic datasets and identified multiple ACE2 protein-altering variants. Using reported structural data, we identified natural ACE2 variants that could potentially affect virus-host interaction and thereby alter host susceptibility. These include variants S19P, I21V, E23K, K26R, T27A, N64K, T92I, Q102P and H378R that were predicted to increase susceptibility, while variants K31R, N33I, H34R, E35K, E37K, D38V, Y50F, N51S, M62V, K68E, F72V, Y83H, G326E, G352V, D355N, Q388L and D509Y were predicted to be protective variants that show decreased binding to S-protein. Using biochemical assays, we confirmed that K31R and E37K had decreased affinity, and K26R and T92I variants showed increased affinity for S-protein when compared to wildtype ACE2. Consistent with this, soluble ACE2 K26R and T92I were more effective in blocking entry of S-protein pseudotyped virus suggesting that ACE2 variants can modulate susceptibility to SARS-CoV-2.
Conflict of interest statement
Employees of ModMab Therapeutics and MedGenome hold shares/options in their respective companies. The remaining authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

