Population genomics and antimicrobial resistance dynamics of Escherichia coli in wastewater and river environments
- PMID: 33846529
- PMCID: PMC8041779
- DOI: 10.1038/s42003-021-01949-x
Population genomics and antimicrobial resistance dynamics of Escherichia coli in wastewater and river environments
Abstract
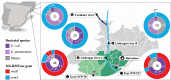
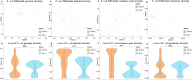
Aquatic environments are key niches for the emergence, evolution and dissemination of antimicrobial resistance. However, the population diversity and the genetic elements that drive the dynamics of resistant bacteria in different aquatic environments are still largely unknown. The aim of this study was to understand the population genomics and evolutionary events of Escherichia coli resistant to clinically important antibiotics including aminoglycosides, in anthropogenic and natural water ecosystems. Here we show that less different E. coli sequence types (STs) are identified in wastewater than in rivers, albeit more resistant to antibiotics, and with significantly more plasmids/cell (6.36 vs 3.72). However, the genomic diversity within E. coli STs in both aquatic environments is similar. Wastewater environments favor the selection of conserved chromosomal structures associated with diverse flexible plasmids, unraveling promiscuous interplasmidic resistance genes flux. On the contrary, the key driver for river E. coli adaptation is a mutable chromosome along with few plasmid types shared between diverse STs harboring a limited resistance gene content.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

