Schlafen 5 as a novel therapeutic target in pancreatic ductal adenocarcinoma
- PMID: 33846574
- PMCID: PMC8106654
- DOI: 10.1038/s41388-021-01761-1
Schlafen 5 as a novel therapeutic target in pancreatic ductal adenocarcinoma
Abstract
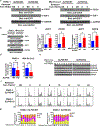
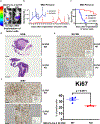
We provide evidence that a member of the human Schlafen (SLFN) family of proteins, SLFN5, is overexpressed in human pancreatic ductal adenocarcinoma (PDAC). Targeted deletion of SLFN5 results in decreased PDAC cell proliferation and suppresses PDAC tumorigenesis in in vivo PDAC models. Importantly, high expression levels of SLFN5 correlate with worse outcomes in PDAC patients, implicating SLFN5 in the pathophysiology of PDAC that leads to poor outcomes. Our studies establish novel regulatory effects of SLFN5 on cell cycle progression through binding/blocking of the transcriptional repressor E2F7, promoting transcription of key genes that stimulate S phase progression. Together, our studies suggest an essential role for SLFN5 in PDAC and support the potential for developing new therapeutic approaches for the treatment of pancreatic cancer through SLFN5 targeting.
Conflict of interest statement
Figures





Similar articles
-
S100A14 promotes progression and gemcitabine resistance in pancreatic cancer.Pancreatology. 2021 Apr;21(3):589-598. doi: 10.1016/j.pan.2021.01.011. Epub 2021 Jan 22. Pancreatology. 2021. PMID: 33579599
-
UBL4A inhibits autophagy-mediated proliferation and metastasis of pancreatic ductal adenocarcinoma via targeting LAMP1.J Exp Clin Cancer Res. 2019 Jul 9;38(1):297. doi: 10.1186/s13046-019-1278-9. J Exp Clin Cancer Res. 2019. PMID: 31288830 Free PMC article.
-
ABCC3 is a novel target for the treatment of pancreatic cancer.Adv Biol Regul. 2019 Aug;73:100634. doi: 10.1016/j.jbior.2019.04.004. Epub 2019 Apr 24. Adv Biol Regul. 2019. PMID: 31053501
-
Central role of Yes-associated protein and WW-domain-containing transcriptional co-activator with PDZ-binding motif in pancreatic cancer development.World J Gastroenterol. 2019 Apr 21;25(15):1797-1816. doi: 10.3748/wjg.v25.i15.1797. World J Gastroenterol. 2019. PMID: 31057295 Free PMC article. Review.
-
Molecular alterations and targeted therapy in pancreatic ductal adenocarcinoma.J Hematol Oncol. 2020 Oct 2;13(1):130. doi: 10.1186/s13045-020-00958-3. J Hematol Oncol. 2020. PMID: 33008426 Free PMC article. Review.
Cited by
-
tRNA-like Transcripts from the NEAT1-MALAT1 Genomic Region Critically Influence Human Innate Immunity and Macrophage Functions.Cells. 2022 Dec 8;11(24):3970. doi: 10.3390/cells11243970. Cells. 2022. PMID: 36552736 Free PMC article.
-
Targeting SLFN11-regulated pathways restores chemotherapy sensitivity in AML.Blood Neoplasia. 2024 Aug 28;1(4):100037. doi: 10.1016/j.bneo.2024.100037. eCollection 2024 Dec. Blood Neoplasia. 2024. PMID: 40552140 Free PMC article.
-
Regulation of IFNα-induced expression of the short ACE2 isoform by ULK1.Mol Immunol. 2022 Jul;147:1-9. doi: 10.1016/j.molimm.2022.04.008. Epub 2022 Apr 27. Mol Immunol. 2022. PMID: 35489289 Free PMC article.
-
SLFN11 negatively regulates non-canonical NFkB signaling to promote glioblastoma progression.Cancer Res Commun. 2022 Sep;2(9):966-978. doi: 10.1158/2767-9764.crc-22-0192. Epub 2022 Sep 13. Cancer Res Commun. 2022. PMID: 36382088 Free PMC article.
-
Progress in investigating the relationship between Schlafen5 genes and malignant tumors.Front Oncol. 2023 Sep 11;13:1248825. doi: 10.3389/fonc.2023.1248825. eCollection 2023. Front Oncol. 2023. PMID: 37771431 Free PMC article. Review.
References
-
- Liu F, Zhou P, Wang Q, Zhang M, Li D. The Schlafen family: complex roles in different cell types and virus replication. Cell Biol Int 2018; 42: 2–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

