How the west was won: genetic reconstruction of rapid wolf recolonization into Germany's anthropogenic landscapes
- PMID: 33846578
- PMCID: PMC8249462
- DOI: 10.1038/s41437-021-00429-6
How the west was won: genetic reconstruction of rapid wolf recolonization into Germany's anthropogenic landscapes
Abstract
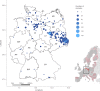
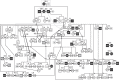
Following massive persecution and eradication, strict legal protection facilitated a successful reestablishment of wolf packs in Germany, which has been ongoing since 2000. Here, we describe this recolonization process by mitochondrial DNA control-region sequencing, microsatellite genotyping and sex identification based on 1341 mostly non-invasively collected samples. We reconstructed the genealogy of German wolf packs between 2005 and 2015 to provide information on trends in genetic diversity, dispersal patterns and pack dynamics during the early expansion process. Our results indicate signs of a founder effect at the start of the recolonization. Genetic diversity in German wolves is moderate compared to other European wolf populations. Although dispersal among packs is male-biased in the sense that females are more philopatric, dispersal distances are similar between males and females once only dispersers are accounted for. Breeding with close relatives is regular and none of the six male wolves originating from the Italian/Alpine population reproduced. However, moderate genetic diversity and inbreeding levels of the recolonizing population are preserved by high sociality, dispersal among packs and several immigration events. Our results demonstrate an ongoing, rapid and natural wolf population expansion in an intensively used cultural landscape in Central Europe.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Andersen LW, Harms V, Caniglia R, Czarnomska SD, Fabbri E, Jędrzejewska B, et al. Long-distance dispersal of a wolf, Canis lupus, in northwestern Europe. Mamm Res. 2015;60:163–168.
-
- Ansorge H, Kluth G, Hahne S. Feeding ecology of wolves Canis lupus returning to Germany. Acta Theriol. 2006;51:99–106.
-
- Caniglia R, Fabbri E, Galaverni M, Milanesi P, Randi E. Noninvasive sampling and genetic variability, pack structure, and dynamics in an expanding wolf population. J Mammal. 2014;95:41–59.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

