Inferring number of populations and changes in connectivity under the n-island model
- PMID: 33846579
- PMCID: PMC8178352
- DOI: 10.1038/s41437-021-00426-9
Inferring number of populations and changes in connectivity under the n-island model
Erratum in
-
Correction: Inferring number of populations and changes in connectivity under the n-island model.Heredity (Edinb). 2022 May;128(5):386. doi: 10.1038/s41437-022-00511-7. Heredity (Edinb). 2022. PMID: 35301466 Free PMC article. No abstract available.
Abstract
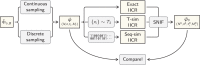
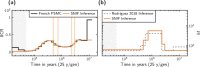
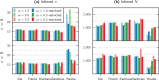
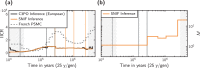
Inferring the demographic history of species is one of the greatest challenges in populations genetics. This history is often represented as a history of size changes, ignoring population structure. Alternatively, when structure is assumed, it is defined a priori as a population tree and not inferred. Here we propose a framework based on the IICR (Inverse Instantaneous Coalescence Rate). The IICR can be estimated for a single diploid individual using the PSMC method of Li and Durbin (2011). For an isolated panmictic population, the IICR matches the population size history, and this is how the PSMC outputs are generally interpreted. However, it is increasingly acknowledged that the IICR is a function of the demographic model and sampling scheme with limited connection to population size changes. Our method fits observed IICR curves of diploid individuals with IICR curves obtained under piecewise stationary symmetrical island models. In our models we assume a fixed number of time periods during which gene flow is constant, but gene flow is allowed to change between time periods. We infer the number of islands, their sizes, the periods at which connectivity changes and the corresponding rates of connectivity. Validation with simulated data showed that the method can accurately recover most of the scenario parameters. Our application to a set of five human PSMCs yielded demographic histories that are in agreement with previous studies using similar methods and with recent research suggesting ancient human structure. They are in contrast with the view of human evolution consisting of one ancestral population branching into three large continental and panmictic populations with varying degrees of connectivity and no population structure within each continent.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

