Genome-wide screens uncover KDM2B as a modifier of protein binding to heparan sulfate
- PMID: 33846619
- PMCID: PMC8159865
- DOI: 10.1038/s41589-021-00776-9
Genome-wide screens uncover KDM2B as a modifier of protein binding to heparan sulfate
Erratum in
-
Author Correction: Genome-wide screens uncover KDM2B as a modifier of protein binding to heparan sulfate.Nat Chem Biol. 2022 May;18(5):575. doi: 10.1038/s41589-022-01022-6. Nat Chem Biol. 2022. PMID: 35354934 No abstract available.
Abstract
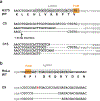
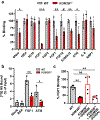
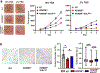
Heparan sulfate (HS) proteoglycans bind extracellular proteins that participate in cell signaling, attachment and endocytosis. These interactions depend on the arrangement of sulfated sugars in the HS chains generated by well-characterized biosynthetic enzymes; however, the regulation of these enzymes is largely unknown. We conducted genome-wide CRISPR-Cas9 screens with a small-molecule ligand that binds to HS. Screening of A375 melanoma cells uncovered additional genes and pathways impacting HS formation. The top hit was the epigenetic factor KDM2B, a histone demethylase. KDM2B inactivation suppressed multiple HS sulfotransferases and upregulated the sulfatase SULF1. These changes differentially affected the interaction of HS-binding proteins. KDM2B-deficient cells displayed decreased growth rates, which was rescued by SULF1 inactivation. In addition, KDM2B deficiency altered the expression of many extracellular matrix genes. Thus, KDM2B controls proliferation of A375 cells through the regulation of HS structure and serves as a master regulator of the extracellular matrix.
Conflict of interest statement
COMPETING INTERESTS
The University of California San Diego and J.D.E. have a financial interest in TEGA Therapeutics, Inc. The terms of this arrangement have been reviewed and approved by the University of California San Diego in accordance with its conflict-of-interest policies.
Figures






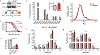

Similar articles
-
Cooperation of binding sites at the hydrophilic domain of cell-surface sulfatase Sulf1 allows for dynamic interaction of the enzyme with its substrate heparan sulfate.Biochim Biophys Acta. 2013 Nov;1830(11):5287-98. doi: 10.1016/j.bbagen.2013.07.014. Epub 2013 Jul 25. Biochim Biophys Acta. 2013. PMID: 23891937
-
TFCP2 is a transcriptional regulator of heparan sulfate assembly and melanoma cell growth.J Biol Chem. 2023 Jun;299(6):104713. doi: 10.1016/j.jbc.2023.104713. Epub 2023 Apr 13. J Biol Chem. 2023. PMID: 37061003 Free PMC article.
-
Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes.Nat Cell Biol. 2013 Apr;15(4):373-84. doi: 10.1038/ncb2702. Epub 2013 Mar 17. Nat Cell Biol. 2013. PMID: 23502314 Free PMC article.
-
Heparan Sulfate Biosynthesis in Zebrafish.J Histochem Cytochem. 2021 Jan;69(1):49-60. doi: 10.1369/0022155420973980. Epub 2020 Nov 20. J Histochem Cytochem. 2021. PMID: 33216642 Free PMC article. Review.
-
Tinkering with heparan sulfate sulfation to steer development.Trends Cell Biol. 2007 Apr;17(4):173-7. doi: 10.1016/j.tcb.2007.02.006. Epub 2007 Feb 21. Trends Cell Biol. 2007. PMID: 17320398 Review.
Cited by
-
B3GALT6 promotes dormant breast cancer cell survival and recurrence by enabling heparan sulfate-mediated FGF signaling.Cancer Cell. 2024 Jan 8;42(1):52-69.e7. doi: 10.1016/j.ccell.2023.11.008. Epub 2023 Dec 7. Cancer Cell. 2024. PMID: 38065100 Free PMC article.
-
Nucleic acid-triggered tumoral immunity propagates pH-selective therapeutic antibodies through tumor-driven epitope spreading.Cancer Sci. 2023 Jan;114(1):321-338. doi: 10.1111/cas.15596. Epub 2022 Oct 17. Cancer Sci. 2023. PMID: 36136061 Free PMC article.
-
Excavating proteoglycan structure-function relationships: modern approaches to capture the interactions of ancient biomolecules.Am J Physiol Cell Physiol. 2022 Aug 1;323(2):C415-C422. doi: 10.1152/ajpcell.00222.2022. Epub 2022 Jun 27. Am J Physiol Cell Physiol. 2022. PMID: 35759439 Free PMC article. Review.
-
Glycosaminoglycans: Participants in Microvascular Coagulation of Sepsis.Thromb Haemost. 2024 Jul;124(7):599-612. doi: 10.1055/a-2250-3166. Epub 2024 Jan 19. Thromb Haemost. 2024. PMID: 38242171 Free PMC article. Review.
-
Reading the glyco-code: New approaches to studying protein-carbohydrate interactions.Curr Opin Struct Biol. 2022 Aug;75:102395. doi: 10.1016/j.sbi.2022.102395. Epub 2022 May 30. Curr Opin Struct Biol. 2022. PMID: 35653954 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

