Mechanisms of microRNA‑142 in mitochondrial autophagy and hippocampal damage in a rat model of epilepsy
- PMID: 33846769
- PMCID: PMC8043661
- DOI: 10.3892/ijmm.2021.4931
Mechanisms of microRNA‑142 in mitochondrial autophagy and hippocampal damage in a rat model of epilepsy
Abstract
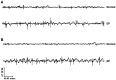
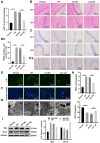
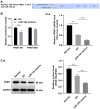
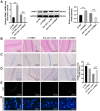
Researchers have confirmed the microRNA (miRNA/miR)‑epilepsy association in rodent models of human epilepsy via a comprehensive database. However, the mechanisms of miR‑142 in epilepsy have not been extensively studied. In the present study, a rat model of epilepsy was first established by an injection of lithium chloride‑pilocarpine and the successful establishment of the model was verified via electroencephalogram monitoring. The levels of miR‑142, phosphatase and tensin homolog deleted on chromosome 10 (PTEN)‑induced putative kinase 1 (PINK1), marker proteins of mitochondrial autophagy, and apoptosis‑related proteins were measured. Additionally, the pathological changes in the hippocampus, the ultrastructure of the mitochondria, and degeneration and the apoptosis of neurons were observed using different staining methods. The malondialdehyde (MDA) content and superoxide dismutase (SOD) activity in the hippocampus, mitochondrial membrane potential (MTP) and reactive oxygen species (ROS) generation were detected. Furthermore, the targeting association between miR‑142 and PINK1 was predicted and verified. Consequently, apoptosis increased, and mitochondrial autophagy decreased, in the hippocampus of epileptic rats. Following miR‑142 inhibition, the epileptic rats exhibited an increased Bax expression, a decreased Bcl‑2 expression, upregulated marker protein levels of mitochondrial autophagy, a reduced MDA content, an enhanced SOD activity, an increased MTP and decreased ROS generation. PINK1 is a target gene of miR‑142, and its overexpression protected against hippocampal damage. Taken together, the results of the present study demonstrated that miR‑142 inhibition promotes mitochondrial autophagy and reduces hippocampal damage in epileptic rats by targeting PINK1. These findings may provide useful information for the treatment of epilepsy.
Keywords: epilepsy; microRNA‑142; PTEN‑induced putative kinase 1; mitochondrial autophagy; hippocampal damage.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

