Inhibition of SARS-CoV-2 main protease: a repurposing study that targets the dimer interface of the protein
- PMID: 33847241
- PMCID: PMC8054500
- DOI: 10.1080/07391102.2021.1910571
Inhibition of SARS-CoV-2 main protease: a repurposing study that targets the dimer interface of the protein
Abstract
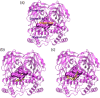
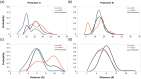
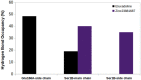
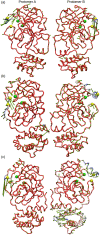
Coronavirus disease-2019 (COVID-19) was firstly reported in Wuhan, China, towards the end of 2019, and emerged as a pandemic. The spread and lethality rates of the COVID-19 have ignited studies that focus on the development of therapeutics for either treatment or prophylaxis purposes. In parallel, drug repurposing studies have also come into prominence. Herein, we aimed at having a holistic understanding of conformational and dynamical changes induced by an experimentally characterized inhibitor on main protease (Mpro) which would enable the discovery of novel inhibitors. To this end, we performed molecular dynamics simulations using crystal structures of apo and α-ketoamide 13b-bound Mpro homodimer. Analysis of trajectories pertaining to apo Mpro revealed a new target site, which is located at the homodimer interface, next to the catalytic dyad. Thereafter, we performed ensemble-based virtual screening by exploiting the ZINC and DrugBank databases and identified three candidate molecules, namely eluxadoline, diosmin, and ZINC02948810 that could invoke local and global conformational rearrangements which were also elicited by α-ketoamide 13b on the catalytic dyad of Mpro. Furthermore, ZINC23881687 stably interacted with catalytically important residues Glu166 and Ser1 and the target site throughout the simulation. However, it gave positive binding energy, presumably, due to displaying higher flexibility that might dominate the entropic term, which is not included in the MM-PBSA method. Finally, ZINC20425029, whose mode of action was different, modulated dynamical properties of catalytically important residue, Ala285. As such, this study presents valuable findings that might be used in the development of novel therapeutics against Mpro.Communicated by Ramaswamy H. Sarma.
Keywords: COVID-19; drug repurposing; main protease; molecular dynamics; novel target site.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures










References
-
- Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindahl, E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1-2, 19–25. 10.1016/j.softx.2015.06.001 - DOI
-
- Arun, K. G., Sharanya, C. S., Abhithaj, J., Francis, D., & Sadasivan, C. (2020). Drug repurposing against SARS-CoV-2 using E-pharmacophore based virtual screening, molecular docking and molecular dynamics with main protease as the target. Journal of Biomolecular Structure & Dynamics, 1–12. https://doi.org/10.1080/07391102.2020.1779819 32571168 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
