Transneuronal Dpr12/DIP-δ interactions facilitate compartmentalized dopaminergic innervation of Drosophila mushroom body axons
- PMID: 33847376
- PMCID: PMC8204868
- DOI: 10.15252/embj.2020105763
Transneuronal Dpr12/DIP-δ interactions facilitate compartmentalized dopaminergic innervation of Drosophila mushroom body axons
Abstract
The mechanisms controlling wiring of neuronal networks are not completely understood. The stereotypic architecture of the Drosophila mushroom body (MB) offers a unique system to study circuit assembly. The adult medial MB γ-lobe is comprised of a long bundle of axons that wire with specific modulatory and output neurons in a tiled manner, defining five distinct zones. We found that the immunoglobulin superfamily protein Dpr12 is cell-autonomously required in γ-neurons for their developmental regrowth into the distal γ4/5 zones, where both Dpr12 and its interacting protein, DIP-δ, are enriched. DIP-δ functions in a subset of dopaminergic neurons that wire with γ-neurons within the γ4/5 zone. During metamorphosis, these dopaminergic projections arrive to the γ4/5 zone prior to γ-axons, suggesting that γ-axons extend through a prepatterned region. Thus, Dpr12/DIP-δ transneuronal interaction is required for γ4/5 zone formation. Our study sheds light onto molecular and cellular mechanisms underlying circuit formation within subcellular resolution.
Keywords: IgSF; circuit formation; dopaminergic neurons; mushroom body compartments; neuronal remodeling.
© 2021 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

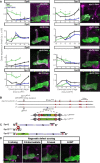
- A
Schematic representation of neuronal remodeling of γ‐KCs and its regulation by the nuclear receptors EcR and UNF. p: axon peduncle; m/v: medial and vertical lobes.
- B
Dynamic expression of Dprs and DIPs during γ‐KC development. Left: Heatmap depicting the relative expression patterns of Dprs and DIPs in γ‐KCs during development. Middle: Magenta intensity depicts the peak expression of each gene during development relative to other Dprs and DIPs. Right: Expression change of Dprs and DIPs while knocking down the UNF transcription factor compared to WT γ‐KCs. Dprs highlighted in bold were tested in the RNAi mini‐screen (Fig EV1).
- C–N
Confocal z‐projections of the indicated genotypes and age, labeled with membrane‐bound GFP (mCD8‐GFP; CD8) driven by the γ‐specific Gal4 driver GMR71G10‐Gal4 (γ‐Gal4). While γ‐axons of control flies project through the entire lobe (C is the RNAi control; n = 12/12, E is the tsCRISPR control; n = 14/14), knockdown of dpr12 by RNAi (D; n = 12/12) or knockout by tsCRISPR (F; n = 14/14) resulted in short axons. At L3, γ‐axons in dpr12∆50‐81 homozygous mutant animals (K; n = 20/20) resemble WT γ‐axons (G; n = 20/20). At 48 h APF, WT γ‐axons normally re‐extend to form the adult lobe (H; n = 12/12). dpr12∆50‐81 γ‐axons (L; n = 14/14) fail to extend to the end of the lobe. This defect persists to adult (I; n = 11/11 vs. M; n = 18/18). Expressing a UAS‐Dpr12 transgene within γ‐KCs in dpr12∆50‐81 homozygote mutant animals rescued the axon regrowth defect (N; n = 23/24, J; n = 14/14). The adult γ‐lobe and α/β lobes are outlined in (C, D) in yellow and orange, respectively, for clarity. Asterisks demarcate the distal part of the lobe. Green and white indicate mCD8‐GFP. Magenta represents FasII staining. Scale bar is 20 µm.
- O
Quantification of the regrowth defects in (I, M, and N). The z‐projections were blindly classified into four classes of regrowth defect severity; see Fig EV1D for examples. Significance was calculated by Kruskal–Wallis test followed by a Mann–Whitney post hoc test; ***P < 0.001.
- P
Models based on hemibrain EM traces (Scheffer et al, 2020) of adult γ‐KCs (representative neurons shown in green), in relation to either selected PAM‐DANs (left; red and orange) or MBONs (right; cyan and blue) targeting the γ4 and γ5 zones. Note that the cell body of the γ4‐MBON is located in the contralateral hemisphere. The MB neuropil is shown in gray. See Materials and Methods for additional details.
- Q
Schematic representation of the adult MB. The bundled γ‐KCs form the γ‐lobe (an example of a single γ‐KC is depicted in green). The γ1‐γ5 zones are defined by stereotyped and tiled innervations of the γ‐lobe by dopaminergic neurons (DANs; examples of DANs targeting the γ4 and γ5 zones are depicted in red and orange, respectively) and MB output neurons (MBONs; an example of the γ4 > γ1γ2 MBON innervation is shown in cyan to match the schematics in Fig 7E and F; note that its cell body and innervations are located in contralateral hemispheres). Black dashed line represents the midline. Magenta represents typical FasII staining (which stains the α/β lobes and the γ‐lobe but not the α’/β’ lobes).
- A
Left: Graphs depicting the normalized RNA expression levels of selected Dprs in WT γ‐KCs (black), and in γ‐KCs expressing EcRDN (green) or UNF‐RNAi (blue). *P < 0.05; Error bars indicate SEM; units on the y‐axis are arbitrary. Right: Confocal z‐projections of adult γ‐KCs expressing RNAi transgenes as indicated labeled with membrane‐bound GFP (mCD8‐GFP; CD8) driven by the γ‐specific Gal4 driver R71G10‐Gal4 (γ‐Gal4). Note that while R71G10 is consistently expressed in γ‐KCs, it is also expressed in α/β‐KCs in a stochastic manner.
- B
A schematic representation of the Dpr12 locus showing introns (black line) and coding and non‐coding exons (red and gray, respectively). The location of Dpr12 gRNA (arrow), dpr12∆50‐81 mutation (arrow), dpr12 RNAiJF03210 (black lines connected with dashed lines), and MiMICMI01695 (arrowhead) is indicated. SA and SD are splice acceptor and donor sites, respectively. Recombination‐mediated cassette exchange was used to transform Dpr12MI01695 into Dpr12GFSTF.
- C
A schematic representation of Dpr12 protein variants. Signal peptide (SP), Immunoglobulin (Ig), transmembrane (TM), and GPI anchor (GPI).
- D
Ranking of regrowth: Confocal z‐projections of adult γ‐KCs labeled with membrane‐bound GFP (mCD8‐GFP; CD8) driven by the γ‐specific Gal4 driver GMR71G10‐Gal4 (γ‐Gal4). Representative images of the regrowth defect severity (1 = strong, 2 = intermediate, 3 = weak, 4 = WT) described in Fig 1O. The arrowhead demarcates an unusually short β‐lobe; morphologically abnormal β‐lobes (either short, thin or absent) appear in approximately 40% of either dpr12 or DIP‐δ homozygous mutant brains. Since β‐lobe morphology is rescued by overexpressing a UAS‐Dpr12 transgene within γKCs, it is most likely a non‐cell‐autonomous defect, which is beyond the scope of this study. Asterisk demarcates the distal tip of the γ‐lobe. Green is CD8‐GFP; magenta is FasII staining. Scale bar is 20 µm.
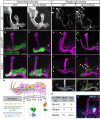
- A–L
Confocal z‐projections of MARCM neuroblast (NB, A‐F) and single‐cell (SC, G‐L) clones labeled with membrane‐bound GFP (mCD8‐GFP; CD8) driven by the γ‐specific Gal4 driver GMR71G10‐Gal4 (γ‐Gal4). At L3, NB and SC clones expressing dpr12 RNAi are similar to equivalent WT clones (A; n = 20/20, D; n = 15/15, G; n = 15/15 and J; n = 17/17). At 48 h APF and adult stage, WT NB (B; n = 15/15, C; n = 10/10) and SC (H; n = 16/16, I; n = 13/13) clones extend their axons to form the full adult lobe. In contrast, clones expressing dpr12 RNAi (E; n = 14/14, F; n = 22/2, K; n = 18/24, L; n = 19/27) fail to extend their axons to the distal part of the medial lobe (asterisks). (I’ and L’) are traces of multiple single‐cell clones depicting each cell in a different color.
- M
Top: Schematic representation of WT (orange) and dpr12 RNAi‐expressing (green) single γ‐KC axons. Bottom: Measurements of the relative location to which WT (I) and dpr12 RNAi (L) axons grow across the entire length of the adult γ lobe, alongside the relative position of the proximal border of the γ4 zone (see O, as well as Fig EV2).
- N
A table depicting the percentage of dpr12 RNAi‐expressing single‐cell clones (SCCs) which stop at the γ3‐γ4 border, at 48 h APF compared to the adult stage.
- O
Confocal z‐projection of MBONγ4 > γ1γ2 labeled by GMR18H09‐Gal4 driving the expression of mCD8‐GFP (CD8) shown in cyan.
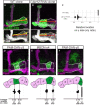
- A, B
Confocal z‐projections of WT (A) and dpr12 RNAi (B) MARCM neuroblast clones (NBC) labeled with membrane‐bound GFP (mCD8‐GFP; CD8) driven by the γ‐specific Gal4 driver GMR71G10‐Gal4 (γ‐Gal4). y (red) represents the length of the entire γ‐lobe as indicated; x (yellow) represents the extent of clonal γ‐axon outgrowth.
- C
Measurements of the x/y ration as depicted in (A, B). While WT NBC always extends up to the end of the lobe, dpr12 RNAi NBC stops at about midway (x/y ratio of 0.48 ± 0.05).
- D–F
Top: Confocal z‐projections of PAM‐DAN‐γ3 (D, MB441B), MBON‐γ4 > γ1γ2 (E, R18H09), and PAM‐DAN‐γ5 (F, R48H11) Gal4s driving the expression of mCD8‐GFP (CD8). Bottom: start and end of the indicated zone is superimposed on a schematic representation of the adult γ lobe compartments. (D) γ3 zone begins at x/y ratio of 0.22 ± 0.03 and ends at 0.48 ± 0.03. (E) γ4 zone begins at x/y ratio of 0.42 ± 0.04 and ends at 0.85 ± 0.03. (F) γ5 zone begins at x/y ratio of 0.76 ± 0.03 and ends at a mean ratio of 1. Green is CD8‐GFP; magenta and white represent FasII. Scale bar is 20 µm.

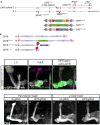
- A–H
Confocal z‐projections of brains expressing MiMIC mediated Dpr12GFSTF (Dpr12‐GFP; A‐D) and DIP‐δGFSTF (DIP‐δ‐GFP; E‐H) fusion proteins at the indicated time points. See Figs EV1 and EV3 for more details on the fusion protein structure. (A‐D) Dpr12‐GFP is localized to the distal part of the γ‐lobe at L3 (A; n = 10/10), 48 h APF (C; n = 12/12) and the adult stage (D; n = 20/20), where it colocalizes with the γ4 > γ1γ2 MBON (γ4; labeled by GMR18H09‐Gal4 driving the expression of CD4‐tdT). At 24hr APF (B; n = 10/10), Dpr12‐GFP appears diffuse. (E‐H) DIP‐δ‐GFP is localized to the distal part of the γ‐lobe throughout development: L3 (E; n = 16/16), 24 h APF (F; n = 10/10), 48 h APF (G; n = 12/12), and adult (H; n = 24/24). At the adult stage, DIP‐δ‐GFP is colocalized with the γ4 > γ1γ2 MBON (γ4).
- A
A schematic representation of the DIP‐δ locus showing introns (black line) and coding and non‐coding exons (red and gray, respectively). The location of DIP‐δ gRNA, DIP‐δ1‐119 mutation, DIP‐δ RNAi, and MiMICMI08287 is indicated. SA and SD are splice acceptor and donor sites, respectively. Recombination‐mediated cassette exchange was used to transform DIP‐δMI08287 into DIP‐δGFSTF and DIP‐δT2A‐Gal4.
- B
A schematic description of DIP‐δ protein variants. Signal peptide (SP), Immunoglobulin (Ig), and GPI anchor (GPI).
- C–E
Confocal z‐projections of DIP‐δ transheterozygotes (DIP‐δT2A‐Gal4/1‐119) at L3 (C; n = 20/20) and adult (D; n = 12/12, E; n = 22/23), in which γ neurons were marked by expressing membrane‐bound tandem tomato (mtdT‐HA) driven by the γ‐specific QF2 driver GMR71G10‐QF2 (γ‐QF2). In DIP‐δ transheterozygotes, as in homozygous mutants (see Fig 4), γ‐KCs do not extend into the distal end of the lobe. Expression of DIP‐δ in DIP‐δ+ cells (E) rescues the growth defect present in DIP‐δT2A‐Gal4/1‐119 brains. Green and white represent mtdT‐HA; magenta represents FasII staining.
- F–I
Confocal z‐projections of brains expressing UAS‐Cas9 alone (F, H) or together with DIP‐δ‐gRNA (G, I). DIP‐δ knockout by tsCRISPR in all postmitotic neurons (G; n = 22/24) resulted in a defect in γ4/5 innervation by γ‐axons, while DIP‐δ knockout by tsCRISPR in γ‐KCs (I; n = 28/28) did not affect γ‐axon regrowth. Expression of Cas9 alone (F, n = 10/10; H, n = 14/14) did not affect γ‐axon regrowth. White represents FasII staining.
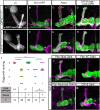
- A–H
Confocal z‐projections DIP‐δ hetero‐ and homozygous brains in which γ‐KCs were labeled by expressing membrane‐bound tandem tomato (mtdT‐HA) driven by the γ‐specific QF2 driver R71G10‐QF2 (γ‐QF2). Larval (L3) γ‐axons grow normally in DIP‐δT2A‐Gal4 heterozygotes (A; n = 16/16) and homozygotes (E; n = 24/24). In contrast, at 48 h APF and adult, γ‐axons within DIP‐δT2A‐Gal4 homozygotes do not enter the distal part of the lobe (asterisks; F; n = 8/8, G; n = 20/20), while they grow normally in heterozygotes (B; n = 12/12, C; n = 12/12). Overexpression of a DIP‐δ transgene driven by the Gal4 activity of DIP‐δT2A‐Gal4 (see also Fig EV3) does not affect normal growth (D; n = 10/10) and rescues mutant phenotypes (H; n = 20/20). Note that while the R71G10 driver is consistently expressed in γ‐KCs, it is also expressed in α/β‐KCs in a stochastic manner. The adult γ‐lobe and α/β lobes are outlined in (C, D) in yellow and orange, respectively, for clarity.
- I
Ranking of regrowth for (C, G, H), and Fig EV3D and E. Regrowth defect severity and statistics were calculated as in Fig 1; Wilcoxon–Mann–Whitney test; ***P < 0.001.
- J–M
Confocal z‐projections of brains expressing DIP‐δ‐RNAi driven by the indicated Gal4. γ‐KCs are labeled by mtdT‐HA driven by R71G10‐QF2 (γ‐QF2). Expression of DIP‐δ‐RNAi in all glia (Repo‐Gal4, J; n = 28/28) or all KCs (OK107‐Gal4, K; n = 12/12) did not affect γ‐neuron regrowth. In contrast, expression of DIP‐δ‐RNAi in all postmitotic neurons (C155‐Gal4, L; n = 9/12) or DIP‐δ‐expressing neurons (DIP‐δT2A‐Gal4, M; n = 21/22) induced a defect in γ4/5 innervation by γ‐axons.
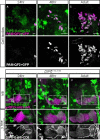
- A–C
Confocal z‐projections of the cell body region of the PAM‐DAN cluster demonstrating the expression of membrane‐bound tandem tomato (CD4‐tdT; CD4) driven by DIP‐δT2A‐Gal4 (DIP‐δ‐Gal4) in addition to GFP driven by the PAM‐DAN specific QF2 driver GMR58E02‐QF2 (PAM‐QF2). The PAM‐QF2 driver is not expressed at 24 h APF (A, n = 12), starts to be expressed at 48 h APF (B, n = 10) and fully expressed and localized with DIP‐δ‐Gal4 in adult (C, n = 14).
- D–F
Confocal z‐projections of heterozygous brains (DIP‐δ+/T2A‐Gal4) in which DIP‐δ positive neurons were labeled by membrane‐bound GFP (mCD8‐GFP; CD8) driven by DIP‐δ‐Gal4. γ‐KCs were marked by expressing membrane‐bound tandem tomato (mtdT‐HA) driven by the γ‐specific QF2 driver GMR71G10‐QF2 (γ‐QF2). Bottom: High magnification images as demarcated by dashed boxes in top panels, of sub‐z‐projections restricted to slices that contain the γ‐lobe.

- A–I
(A, B, D, E, G, H) Left: Detailed analysis of the confocal z‐projections of dpr12 heterozygous or homozygous mutant brains that are presented in Fig 7A–F, which express membrane‐bound GFP (CD8) driven by: (A‐B) R10G03‐Gal4 is used to label PAM‐DANs innervating the γ4 compartment (PAM‐DAN‐γ4); (D‐E) R48H11‐Gal4 is used to label PAM‐DANs innervating the γ5 compartment (PAM‐DAN‐γ5); (G‐H) R18H09‐Gal4 is used to label the MBONγ4 > γ1γ2 which innervates the γ4 zone (MBON‐γ4). Right: YZ projections along the indicated lines in γ3 (orange) and γ4 or γ5 (blue) compartments. (C, F, I) Top: Cell body numbers of the indicated neurons in dpr12 heterozygous and homozygous brains. Bottom: Summary of innervation destinations. Unknown means stereotypic projections to unidentifiable domains.
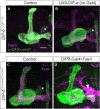
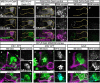
- A, B
Confocal z‐projections of DIP‐δ1‐119 homozygous mutant brains, in which γ‐KCs are labeled by membrane‐bound tandem tomato (mtdT‐HA; green) driven by R71G10‐QF2 (γ‐QF2), that either contain (B) or do not contain (A) a UAS‐DIP‐α transgene.
- C, D
Confocal z‐projections of DIP‐δT2A‐Gal4/T2A‐Gal4 homozygous mutant brains, in which γ‐KCs are labeled by mtdT‐HA (green) driven by γ‐QF2, that either express (D) or do not express (C) a UAS‐FasII transgene driven by DIP‐δ‐Gal4.
- A–D
Confocal z‐projections of brains expressing DIP‐δGFSTF (DIP‐δ‐GFP) together with the indicated Gal4s and transgenes. Expressing diphtheria toxin (DTi) and membrane‐bound RFP (mCD8‐RFP; CD8) driven by the γ4 > γ1γ2 MBON driver MB294B‐Gal4 (MBONγ4‐Gal4) did not affect DIP‐δ‐GFP expression (A, n = 16/16; B, n = 14/14). In contrast, similar expression of DTi and membrane‐bound tomato (CD4‐tdT; CD4) in PAM‐DANs (using the GMR58E02‐Gal4; PAM‐DAN‐Gal4) abolished the normal DIP‐δ‐GFP expression in the γ4/5 zone (compare D, n = 18/18, to C, n = 16/16) and within the PAM‐DAN cell bodies (compare D’ to C’). Magenta is CD8‐RFP (A, B) and CD4‐mtdT (C, D). Green is GFP, and grayscale depicts individual channels as labeled. Scale bar is 20 µm. Yellow dashed line demarcates the γ‐lobe based on FasII staining (not shown).
- E–G
Confocal z‐projections of MARCM clones labeled by DIP‐δT2A‐Gal4 (DIP‐δ Gal4) driving the expression of membrane‐bound GFP (mCD8‐GFP; CD8) and heat shocked at 24 h after egg laying. Clones innervate the γ4/5 zones at 24 h APF (E; n = 8), 48 h APF (F; n = 8), and adult (G; n = 16). Clones become tyrosine hydroxylase (TH) positive only at 48 h APF onwards (F, G). Magenta is FasII, green is mCD8‐GFP, cyan is TH antibody staining, and grayscale single channels are shown as indicated. White dashed line demarcates the γ‐lobe. Scale bar is 20 µm.
- A–J
Confocal z‐projections of brains expressing MiMIC mediated Dpr12GFSTF (Dpr12‐GFP) and DIP‐δGFSTF (DIP‐δ‐GFP) fusion proteins of the indicated genotypes and time points. (A‐D) Dpr12‐GFP expression is diffuse in DIP‐δT2A‐Gal4 homozygotes mutant brains at L3 (A; n = 20/20), 24 h APF (B; n = 14/14), 48 h APF (C; n = 28/28), and adult (D; n = 26/26). (E‐H) DIP‐δ‐GFP expression in dpr12∆50‐81 homozygotes mutant brains remains localized to the distal part of the γ‐lobe at L3 (E; n = 16/16), 24 h APF (F; n = 16/16), and 48 h APF (G; n = 10/10) but cannot be identified in adult brains (H; n = 16/16). (I, J) Dpr12‐GFP expression in WT animals (I, n = 8/8) or in those ectopically expressing DIP‐δ in PAM‐DANs that innervate the γ3 zone (J, n = 14/14) driven by MB441B‐Gal4 (PAM‐DAN‐γ3‐Gal4). DIP‐δ expression in PAM‐DAN‐γ3 resulted in Dpr12‐GFP localization within the γ3 zone (arrow), in addition to its normal γ4/γ5 localization.

- A–F
Top: Confocal z‐projections of dpr12∆50‐81 heterozygous (A, n = 15; C, n = 10; E, n = 10) and homozygous brains (B, n = 8; D, n = 18; F, n = 24) expressing mCD8‐GFP (CD8) driven by: (A, B) R10G03‐Gal4 (PAM‐DAN‐γ4‐Gal4); (C, D) R48H11‐Gal4 (PAM‐DAN‐γ5‐Gal4), or (E, F) R18H09‐Gal4 (MBONγ4 > γ1γ2 ‐Gal4). The grayscale channels are sub‐z‐projections comprised of slices restricted to the γ‐lobe region. White dashed line demarcates the γ‐lobe. Bottom: Cartoons schematizing MB lobe structure and innervation by specific PAM‐DANs or MBON.
- G, H
Single confocal slices of WT (G, n = 5/5) and dpr12∆50‐81 homozygous brains (H, n = 5/5) stained with anti‐Brp. Dashed lines demarcate neuropil boundaries, schematic shown below. Cre, crepine, a neuropil that surrounds the medial MB lobes; AL, antenna lobe.

- A, B
Confocal z‐projections of adult DIP‐δT2A‐Gal4/ 1‐119 trans‐heterozygous mutant brains, in which γ‐KCs are labeled by membrane‐bound tandem tomato (mtdT‐HA; green) driven by R71G10‐QF2 (γ‐QF2), and DIP‐δ‐Gal4 (expressed in DIP‐δ+ neurons) either drives expression of UAS‐DIP‐α (B) or not (A). Gray boxes below the images describe the relevant components within PAM‐DANs and γ‐KCs.
- C, D
Confocal z‐projections of adult DIP‐δT2A‐Gal4/ 1‐119 trans‐heterozygous mutant (D) or DIP‐δ1‐119/ + heterozygous (C) brains, which express MiMIC mediated Dpr12GFSTF (Dpr12‐GFP; green). In (D), DIP‐δ‐Gal4 drives expression of UAS‐DIP‐α. Grayscale panels represent single channels, as indicated. The γ‐lobe is outlined in white.
- E, F
Confocal z‐projections of adult dpr12Δ50‐81 homozygous mutant brains, in which DIP‐δ‐Gal4 drives expression of either UAS‐mtdT (E) or UAS‐DIP‐α‐T2A‐tdT (F; expected to induce expression of DIP‐α as well as tdT encoded by a polycistronic message). In orange are longitudinal sections across the γ‐lobe at the indicated location. The γ‐lobe is outlined in white, as determined by FasII staining (gray in E’‐F’). Cyan is tdT within DIP‐δ+ cells. Gray boxes below the images describe the relevant components within PAM‐DANs and γ‐KCs.
- G
Ranking of regrowth for (A, B, E, F) and Fig EV6A and B. Regrowth defect severity and statistics were calculated as in Fig 1; Wilcoxon–Mann–Whitney test; ***P < 0.001; ns, not significant.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

