A Natural Product Chemist's Guide to Unlocking Silent Biosynthetic Gene Clusters
- PMID: 33848426
- PMCID: PMC9148385
- DOI: 10.1146/annurev-biochem-081420-102432
A Natural Product Chemist's Guide to Unlocking Silent Biosynthetic Gene Clusters
Abstract
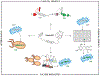
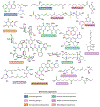
Microbial natural products have provided an important source of therapeutic leads and motivated research and innovation in diverse scientific disciplines. In recent years, it has become evident that bacteria harbor a large, hidden reservoir of potential natural products in the form of silent or cryptic biosynthetic gene clusters (BGCs). These can be readily identified in microbial genome sequences but do not give rise to detectable levels of a natural product. Herein, we provide a useful organizational framework for the various methods that have been implemented for interrogating silent BGCs. We divide all available approaches into four categories. The first three are endogenous strategies that utilize the native host in conjunction with classical genetics, chemical genetics, or different culture modalities. The last category comprises expression of the entire BGC in a heterologous host. For each category, we describe the rationale, recent applications, and associated advantages and limitations.
Keywords: bacteria; biosynthesis; cryptic metabolite; natural product; silent gene cluster.
Figures


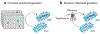
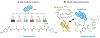

References
-
- Newman DJ, Cragg GM. 2020. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod 83:770–803 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

