Impact of chromatin context on Cas9-induced DNA double-strand break repair pathway balance
- PMID: 33848455
- PMCID: PMC8153251
- DOI: 10.1016/j.molcel.2021.03.032
Impact of chromatin context on Cas9-induced DNA double-strand break repair pathway balance
Abstract
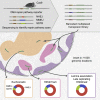
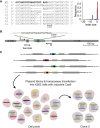
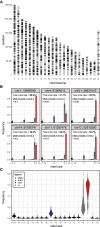
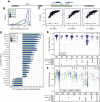
DNA double-strand break (DSB) repair is mediated by multiple pathways. It is thought that the local chromatin context affects the pathway choice, but the underlying principles are poorly understood. Using a multiplexed reporter assay in combination with Cas9 cutting, we systematically measure the relative activities of three DSB repair pathways as a function of chromatin context in >1,000 genomic locations. This reveals that non-homologous end-joining (NHEJ) is broadly biased toward euchromatin, while the contribution of microhomology-mediated end-joining (MMEJ) is higher in specific heterochromatin contexts. In H3K27me3-marked heterochromatin, inhibition of the H3K27 methyltransferase EZH2 reverts the balance toward NHEJ. Single-stranded template repair (SSTR), often used for precise CRISPR editing, competes with MMEJ and is moderately linked to chromatin context. These results provide insight into the impact of chromatin on DSB repair pathway balance and guidance for the design of Cas9-mediated genome editing experiments.
Keywords: CRISPR; Chromatin; DNA repair; MMEJ; NHEJ; SSTR; double strand break; heterochromatin; nuclear lamina; reporter assay.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.v.S. is a member of the Advisory Board of Molecular Cell.
Figures



Comment in
-
Chromatin context affects DNA repair pathway.Nat Rev Genet. 2021 Jul;22(7):414. doi: 10.1038/s41576-021-00371-7. Nat Rev Genet. 2021. PMID: 33963357 No abstract available.
-
A molecular Rosetta Stone to decipher the impact of chromatin features on the repair of Cas9-mediated DNA double-strand breaks.Mol Cell. 2021 May 20;81(10):2059-2060. doi: 10.1016/j.molcel.2021.04.024. Mol Cell. 2021. PMID: 34019786
References
-
- Akhtar W., de Jong J., Pindyurin A.V., Pagie L., Meuleman W., de Ridder J., Berns A., Wessels L.F., van Lohuizen M., van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. - PubMed
-
- Akhtar W., Pindyurin A.V., de Jong J., Pagie L., Ten Hoeve J., Berns A., Wessels L.F., van Steensel B., van Lohuizen M. Using TRIP for genome-wide position effect analysis in cultured cells. Nat. Protoc. 2014;9:1255–1281. - PubMed
-
- Aho A.V., Kernighan B.W., Weinberger P.J. The awk programming language. Comput. Hum. 1992;26:293–297.
-
- Allen F., Crepaldi L., Alsinet C., Strong A.J., Kleshchevnikov V., De Angeli P., Palenikova P., Khodak A., Kiselev V., Kosicki M. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 2018 doi: 10.1038/nbt.4317. Published online November 27, 2018. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

