Nanomechanics of G-quadruplexes within the promoter of the KIT oncogene
- PMID: 33849064
- PMCID: PMC8096272
- DOI: 10.1093/nar/gkab079
Nanomechanics of G-quadruplexes within the promoter of the KIT oncogene
Abstract
G-quadruplexes (G4s) are tetrahelical DNA structures stabilized by four guanines paired via Hoogsteen hydrogen bonds into quartets. While their presence within eukaryotic DNA is known to play a key role in regulatory processes, their functional mechanisms are still under investigation. In the present work, we analysed the nanomechanical properties of three G4s present within the promoter of the KIT proto-oncogene from a single-molecule point of view through the use of magnetic tweezers (MTs). The study of DNA extension fluctuations under negative supercoiling allowed us to identify a characteristic fingerprint of G4 folding. We further analysed the energetic contribution of G4 to the double-strand denaturation process in the presence of negative supercoiling, and we observed a reduction in the energy required for strands separation.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



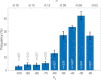
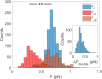

References
-
- Lightfoot H.L., Hagen T., Tatum N.J., Hall J.. The diverse structural landscape of quadruplexes. FEBS Lett. 2019; 593:2083–2102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

