Bimodal regulation of the PRC2 complex by USP7 underlies tumorigenesis
- PMID: 33849069
- PMCID: PMC8096222
- DOI: 10.1093/nar/gkab209
Bimodal regulation of the PRC2 complex by USP7 underlies tumorigenesis
Abstract
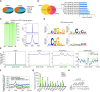
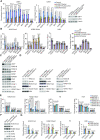
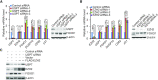
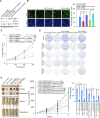
Although overexpression of EZH2, a catalytic subunit of the polycomb repressive complex 2 (PRC2), is an eminent feature of various cancers, the regulation of its abundance and function remains insufficiently understood. We report here that the PRC2 complex is physically associated with ubiquitin-specific protease USP7 in cancer cells where USP7 acts to deubiquitinate and stabilize EZH2. Interestingly, we found that USP7-catalyzed H2BK120ub1 deubiquitination is a prerequisite for chromatin loading of PRC2 thus H3K27 trimethylation, and this process is not affected by H2AK119 ubiquitination catalyzed by PRC1. Genome-wide analysis of the transcriptional targets of the USP7/PRC2 complex identified a cohort of genes including FOXO1 that are involved in cell growth and proliferation. We demonstrated that the USP7/PRC2 complex drives cancer cell proliferation and tumorigenesis in vitro and in vivo. We showed that the expression of both USP7 and EZH2 elevates during tumor progression, corresponding to a diminished FOXO1 expression, and the level of the expression of USP7 and EZH2 strongly correlates with histological grades and prognosis of tumor patients. These results reveal a dual role for USP7 in the regulation of the abundance and function of EZH2, supporting the pursuit of USP7 as a therapeutic target for cancer intervention.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Structural Basis of the Interaction Between Ubiquitin Specific Protease 7 and Enhancer of Zeste Homolog 2.J Mol Biol. 2020 Feb 14;432(4):897-912. doi: 10.1016/j.jmb.2019.12.026. Epub 2019 Dec 20. J Mol Biol. 2020. PMID: 31866294
-
A feedforward circuit shaped by ECT2 and USP7 contributes to breast carcinogenesis.Theranostics. 2020 Aug 29;10(23):10769-10790. doi: 10.7150/thno.46878. eCollection 2020. Theranostics. 2020. PMID: 32929379 Free PMC article.
-
Dual Inhibition of EZH2 and EZH1 Sensitizes PRC2-Dependent Tumors to Proteasome Inhibition.Clin Cancer Res. 2017 Aug 15;23(16):4817-4830. doi: 10.1158/1078-0432.CCR-16-2735. Epub 2017 May 10. Clin Cancer Res. 2017. PMID: 28490465 Free PMC article.
-
Small Molecule Approaches for Targeting the Polycomb Repressive Complex 2 (PRC2) in Cancer.J Med Chem. 2020 Dec 24;63(24):15344-15370. doi: 10.1021/acs.jmedchem.0c01344. Epub 2020 Dec 7. J Med Chem. 2020. PMID: 33283516 Review.
-
Potential of enhancer of zeste homolog 2 inhibitors for the treatment of SWI/SNF mutant cancers and tumor microenvironment modulation.Drug Dev Res. 2021 Sep;82(6):730-753. doi: 10.1002/ddr.21796. Epub 2021 Feb 9. Drug Dev Res. 2021. PMID: 33565092 Review.
Cited by
-
Machine-learning-optimized Cas12a barcoding enables the recovery of single-cell lineages and transcriptional profiles.Mol Cell. 2022 Aug 18;82(16):3103-3118.e8. doi: 10.1016/j.molcel.2022.06.001. Epub 2022 Jun 24. Mol Cell. 2022. PMID: 35752172 Free PMC article.
-
USP7 deubiquitinates and stabilizes EZH2 in prostate cancer cells.Genet Mol Biol. 2020 May 20;43(2):e20190338. doi: 10.1590/1678-4685-GMB-2019-0338. eCollection 2020. Genet Mol Biol. 2020. PMID: 32453339 Free PMC article.
-
The Role and Mechanism of Deubiquitinase USP7 in Tumor-Associated Inflammation.Biomedicines. 2024 Nov 29;12(12):2734. doi: 10.3390/biomedicines12122734. Biomedicines. 2024. PMID: 39767641 Free PMC article. Review.
-
Role of USP7 in the regulation of tolerogenic dendritic cell function in type 1 diabetes.Cell Mol Biol Lett. 2025 Apr 17;30(1):47. doi: 10.1186/s11658-025-00727-5. Cell Mol Biol Lett. 2025. PMID: 40247205 Free PMC article.
-
Synergistic regulation of DACH1 stability by acetylation and deubiquitination promotes colorectal cancer progression.Cell Death Dis. 2025 May 19;16(1):400. doi: 10.1038/s41419-025-07696-9. Cell Death Dis. 2025. PMID: 40389405 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

