Ageing genetic signature of hypersomatotropism
- PMID: 33849304
- PMCID: PMC8059514
- DOI: 10.1098/rsob.200265
Ageing genetic signature of hypersomatotropism
Abstract
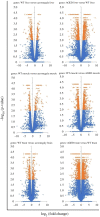
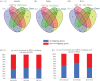
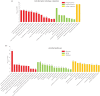
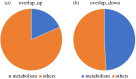
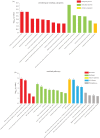
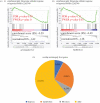
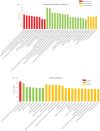
Acromegaly is a pathological condition that is caused by over-secretion of growth hormone (GH) and develops primarily from a pituitary adenoma. Excess GH exposure over a prolonged period of time leads to a wide range of systemic manifestations and comorbidities. Studying the effect of excess GH on the cellular level could help to understand the underlying causes of acromegaly health complications and comorbidities. In our previous publications, we have shown that excess GH reduces body side population (SP) stem cells and induces signs of premature ageing in an acromegaly zebrafish model. Here, we study acromegaly ageing in greater depth at the level of gene expression. We investigated whether acromegaly induces an ageing genetic signature in different organs. Using the GenAge database, our acromegaly model showed a significant enrichment of ageing genetic datasets in the muscle but not in other organs. Likewise, the hierarchical clustering of wild type (WT), acromegaly and aged RNA data from various organs revealed the similarity of gene expression profiles between the acromegaly and the aged muscles. We therefore identified overlapping differentially expressed genes (DEGs) in different organs between acromegaly and aged zebrafish. Importantly, about half of the muscle, liver and brain acromegaly DEGs overlapped with aged zebrafish DEGs. Interestingly, overlapping was observed in the same way; acromegaly-up DEGs overlapped with aged zebrafish up DEGs, not down DEGs, and vice versa. We then identified the biological functions of overlapping DEGs. Enrichment database analysis and gene ontology showed that most overlapping muscle genes were involved in ageing metabolism, while overlapping liver DEGs were involved in metabolic pathways, response to hypoxia and endoplasmic reticulum stress. Thus, this study provides a full ageing genetic signature of acromegaly at the gene expression level.
Keywords: acromegaly; ageing signature; growth hormone; zebrafish.
Figures


Similar articles
-
Induction of endoplasmic reticulum stress markers in an acromegaly model.J Cell Biochem. 2022 Sep;123(9):1411-1421. doi: 10.1002/jcb.30301. Epub 2022 Jun 23. J Cell Biochem. 2022. PMID: 35734917
-
An Acromegaly Disease Zebrafish Model Reveals Decline in Body Stem Cell Number along with Signs of Premature Aging.Biology (Basel). 2020 Jun 7;9(6):120. doi: 10.3390/biology9060120. Biology (Basel). 2020. PMID: 32517323 Free PMC article.
-
A Zebrafish Acromegaly Model Elevates DNA Damage and Impairs DNA Repair Pathways.Biology (Basel). 2018 Oct 17;7(4):47. doi: 10.3390/biology7040047. Biology (Basel). 2018. PMID: 30336646 Free PMC article.
-
Hormonal diagnosis of GH hypersecretory states.J Endocrinol Invest. 2003;26(10 Suppl):27-35. J Endocrinol Invest. 2003. PMID: 15497657 Review.
-
Acromégalie : améliorer la prise en charge: Acromegaly: improving care.Ann Endocrinol (Paris). 2019 Sep;80 Suppl 1:S10-S18. doi: 10.1016/S0003-4266(19)30112-X. Ann Endocrinol (Paris). 2019. PMID: 31606057 Review. French.
References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

