Explore the gene network regulating the composition of fatty acids in cottonseed
- PMID: 33849439
- PMCID: PMC8042725
- DOI: 10.1186/s12870-021-02952-4
Explore the gene network regulating the composition of fatty acids in cottonseed
Abstract
Background: Cottonseed is one of the major sources of vegetable oil. Analysis of the dynamic changes of fatty acid components and the genes regulating the composition of fatty acids of cottonseed oil is of great significance for understanding the biological processes underlying biosynthesis of fatty acids and for genetic improving the oil nutritional qualities.
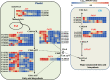
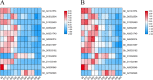
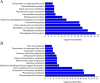
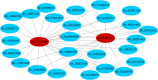
Results: In this study, we investigated the dynamic relationship of 13 fatty acid components at 12 developmental time points of cottonseed (Gossypium hirsutum L.) and generated cottonseed transcriptome of the 12 time points. At 5-15 day post anthesis (DPA), the contents of polyunsaturated linolenic acid (C18:3n-3) and saturated stearic acid (C18:0) were higher, while linoleic acid (C18:2n-6) was mainly synthesized after 15 DPA. Using 5 DPA as a reference, 15,647 non-redundant differentially expressed genes were identified in 10-60 DPA cottonseed. Co-expression gene network analysis identified six modules containing 3275 genes significantly associated with middle-late seed developmental stages and enriched with genes related to the linoleic acid metabolic pathway and α-linolenic acid metabolism. Genes (Gh_D03G0588 and Gh_A02G1788) encoding stearoyl-ACP desaturase were identified as hub genes and significantly up-regulated at 25 DPA. They seemed to play a decisive role in determining the ratio of saturated fatty acids to unsaturated fatty acids. FAD2 genes (Gh_A13G1850 and Gh_D13G2238) were highly expressed at 25-50 DPA, eventually leading to the high content of C18:2n-6 in cottonseed. The content of C18:3n-3 was significantly decreased from 5 DPA (7.44%) to 25 DPA (0.11%) and correlated with the expression characteristics of Gh_A09G0848 and Gh_D09G0870.
Conclusions: These results contribute to our understanding on the relationship between the accumulation pattern of fatty acid components and the expression characteristics of key genes involved in fatty acid biosynthesis during the entire period of cottonseed development.
Keywords: Co-expression network; DEGs; Fatty acid components; Gossypium hirsutum; Transcriptomic analysis.
Conflict of interest statement
The authors report no declarations of interest.
Figures


References
-
- Orsavova J, Misurcova L, Ambrozova JV, Vicha R, Mlcek J. Fatty acids composition of vegetable oils and its contribution to dietary energy intake and dependence of cardiovascular mortality on dietary intake of fatty acids. Int J Mol Sci. 2015;16(6):12871–12890. doi: 10.3390/ijms160612871. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

