Rac1 activation can generate untemplated, lamellar membrane ruffles
- PMID: 33849538
- PMCID: PMC8042924
- DOI: 10.1186/s12915-021-00997-3
Rac1 activation can generate untemplated, lamellar membrane ruffles
Abstract
Background: Membrane protrusions that occur on the dorsal surface of a cell are an excellent experimental system to study actin machinery at work in a living cell. Small GTPase Rac1 controls the membrane protrusions that form and encapsulate extracellular volumes to perform pinocytic or phagocytic functions.
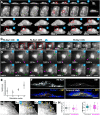
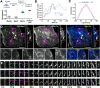
Results: Here, capitalizing on rapid volumetric imaging capabilities of lattice light-sheet microscopy (LLSM), we describe optogenetic approaches using photoactivable Rac1 (PA-Rac1) for controlled ruffle generation. We demonstrate that PA-Rac1 activation needs to be continuous, suggesting a threshold local concentration for sustained actin polymerization leading to ruffling. We show that Rac1 activation leads to actin assembly at the dorsal surface of the cell membrane that result in sheet-like protrusion formation without any requirement of a template. Further, this approach can be used to study the complex morpho-dynamics of the protrusions or to investigate specific proteins that may be enriched in the ruffles. Deactivating PA-Rac1 leads to complex contractile processes resulting in formation of macropinosomes. Using multicolour imaging in combination with these approaches, we find that Myo1e specifically is enriched in the ruffles.
Conclusions: Combining LLSM and optogenetics enables superior spatial and temporal control for studying such dynamic mechanisms. Demonstrated here, the techniques implemented provide insight into the complex nature of the molecular interplay involved in dynamic actin machinery, revealing that Rac1 activation can generate untemplated, lamellar protrusions.
Keywords: Actin polymerization; Lamellipodia; Lattice light-sheet microscopy; Optogenetics; Rac1.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

