mTORC2 controls the activity of PKC and Akt by phosphorylating a conserved TOR interaction motif
- PMID: 33850054
- PMCID: PMC8208635
- DOI: 10.1126/scisignal.abe4509
mTORC2 controls the activity of PKC and Akt by phosphorylating a conserved TOR interaction motif
Abstract
The complex mTORC2 is accepted to be the kinase that controls the phosphorylation of the hydrophobic motif, a key regulatory switch for AGC kinases, although whether mTOR directly phosphorylates this motif remains controversial. Here, we identified an mTOR-mediated phosphorylation site that we termed the TOR interaction motif (TIM; F-x3-F-pT), which controls the phosphorylation of the hydrophobic motif of PKC and Akt and the activity of these kinases. The TIM is invariant in mTORC2-dependent AGC kinases, is evolutionarily conserved, and coevolved with mTORC2 components. Mutation of this motif in Akt1 and PKCβII abolished cellular kinase activity by impairing activation loop and hydrophobic motif phosphorylation. mTORC2 directly phosphorylated the PKC TIM in vitro, and this phosphorylation event was detected in mouse brain. Overexpression of PDK1 in mTORC2-deficient cells rescued hydrophobic motif phosphorylation of PKC and Akt by a mechanism dependent on their intrinsic catalytic activity, revealing that mTORC2 facilitates the PDK1 phosphorylation step, which, in turn, enables autophosphorylation. Structural analysis revealed that PKC homodimerization is driven by a TIM-containing helix, and biophysical proximity assays showed that newly synthesized, unphosphorylated PKC dimerizes in cells. Furthermore, disruption of the dimer interface by stapled peptides promoted hydrophobic motif phosphorylation. Our data support a model in which mTORC2 relieves nascent PKC dimerization through TIM phosphorylation, recruiting PDK1 to phosphorylate the activation loop and triggering intramolecular hydrophobic motif autophosphorylation. Identification of TIM phosphorylation and its role in the regulation of PKC provides the basis for AGC kinase regulation by mTORC2.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
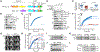

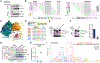

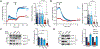


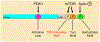
References
-
- Nolen B, Taylor S, Ghosh G, Regulation of protein kinases: Controlling activity through activation segment conformation. Mol. Cell 15 (2004), pp. 661–675. - PubMed
-
- Leroux AE, Schulze JO, Biondi RM, AGC kinases, mechanisms of regulation and innovative drug development. Semin. Cancer Biol 48, 1–17 (2018). - PubMed
-
- Mora A, Komander D, Van Aalten DMF, Alessi DR, PDK1, the master regulator of AGC kinase signal transduction. Semin. Cell Dev. Biol 15, 161–170 (2004). - PubMed
-
- Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PRJ, Reese CB, Cohen P, Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol 7, 261–269 (1997). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 CA239106/CA/NCI NIH HHS/United States
- R01 GM114409/GM/NIGMS NIH HHS/United States
- T32 AR064194/AR/NIAMS NIH HHS/United States
- R01 GM134168/GM/NIGMS NIH HHS/United States
- T32 HL007444/HL/NHLBI NIH HHS/United States
- R37 GM043154/GM/NIGMS NIH HHS/United States
- R01 HL155826/HL/NHLBI NIH HHS/United States
- R01 GM043154/GM/NIGMS NIH HHS/United States
- P 30441/FWF_/Austrian Science Fund FWF/Austria
- R35 GM122523/GM/NIGMS NIH HHS/United States
- T32 GM007752/GM/NIGMS NIH HHS/United States
- R35 GM130389/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

