Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney
- PMID: 33850129
- PMCID: PMC8044133
- DOI: 10.1038/s41467-021-22368-w
Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney
Abstract
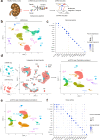
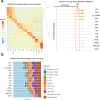
The integration of single cell transcriptome and chromatin accessibility datasets enables a deeper understanding of cell heterogeneity. We performed single nucleus ATAC (snATAC-seq) and RNA (snRNA-seq) sequencing to generate paired, cell-type-specific chromatin accessibility and transcriptional profiles of the adult human kidney. We demonstrate that snATAC-seq is comparable to snRNA-seq in the assignment of cell identity and can further refine our understanding of functional heterogeneity in the nephron. The majority of differentially accessible chromatin regions are localized to promoters and a significant proportion are closely associated with differentially expressed genes. Cell-type-specific enrichment of transcription factor binding motifs implicates the activation of NF-κB that promotes VCAM1 expression and drives transition between a subpopulation of proximal tubule epithelial cells. Our multi-omics approach improves the ability to detect unique cell states within the kidney and redefines cellular heterogeneity in the proximal tubule and thick ascending limb.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

