This is a preprint.
Mendelian randomisation identifies alternative splicing of the FAS death receptor as a mediator of severe COVID-19
- PMID: 33851187
- PMCID: PMC8043484
- DOI: 10.1101/2021.04.01.21254789
Mendelian randomisation identifies alternative splicing of the FAS death receptor as a mediator of severe COVID-19
Abstract
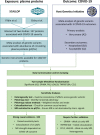
Severe COVID-19 is characterised by immunopathology and epithelial injury. Proteomic studies have identified circulating proteins that are biomarkers of severe COVID-19, but cannot distinguish correlation from causation. To address this, we performed Mendelian randomisation (MR) to identify proteins that mediate severe COVID-19. Using protein quantitative trait loci (pQTL) data from the SCALLOP consortium, involving meta-analysis of up to 26,494 individuals, and COVID-19 genome-wide association data from the Host Genetics Initiative, we performed MR for 157 COVID-19 severity protein biomarkers. We identified significant MR results for five proteins: FAS, TNFRSF10A, CCL2, EPHB4 and LGALS9. Further evaluation of these candidates using sensitivity analyses and colocalization testing provided strong evidence to implicate the apoptosis-associated cytokine receptor FAS as a causal mediator of severe COVID-19. This effect was specific to severe disease. Using RNA-seq data from 4,778 individuals, we demonstrate that the pQTL at the FAS locus results from genetically influenced alternate splicing causing skipping of exon 6. We show that the risk allele for very severe COVID-19 increases the proportion of transcripts lacking exon 6, and thereby increases soluble FAS. Soluble FAS acts as a decoy receptor for FAS-ligand, inhibiting apoptosis induced through membrane-bound FAS. In summary, we demonstrate a novel genetic mechanism that contributes to risk of severe of COVID-19, highlighting a pathway that may be a promising therapeutic target.
Figures


Similar articles
-
A proteome-wide genetic investigation identifies several SARS-CoV-2-exploited host targets of clinical relevance.Elife. 2021 Aug 17;10:e69719. doi: 10.7554/eLife.69719. Elife. 2021. PMID: 34402426 Free PMC article.
-
Biomarkers Associated With Severe COVID-19 Among Populations With High Cardiometabolic Risk: A 2-Sample Mendelian Randomization Study.JAMA Netw Open. 2023 Jul 3;6(7):e2325914. doi: 10.1001/jamanetworkopen.2023.25914. JAMA Netw Open. 2023. PMID: 37498601 Free PMC article.
-
An Integrative Genomic Strategy Identifies sRAGE as a Causal and Protective Biomarker of Lung Function.Chest. 2022 Jan;161(1):76-84. doi: 10.1016/j.chest.2021.06.053. Epub 2021 Jul 6. Chest. 2022. PMID: 34237330 Free PMC article.
-
Mendelian randomization analysis identified tumor necrosis factor as being associated with severe COVID-19.Front Pharmacol. 2023 Jun 16;14:1171404. doi: 10.3389/fphar.2023.1171404. eCollection 2023. Front Pharmacol. 2023. PMID: 37397483 Free PMC article.
-
PTGES2 and RNASET2 identified as novel potential biomarkers and therapeutic targets for basal cell carcinoma: insights from proteome-wide mendelian randomization, colocalization, and MR-PheWAS analyses.Front Pharmacol. 2024 Jul 5;15:1418560. doi: 10.3389/fphar.2024.1418560. eCollection 2024. Front Pharmacol. 2024. PMID: 39035989 Free PMC article.
References
-
- Interleukin-6 Receptor Antagonists in Critically Ill Patients with Covid-19. New England Journal of Medicine (2021). - PubMed
Publication types
Grants and funding
- MC_UU_00007/10/MRC_/Medical Research Council/United Kingdom
- 75N92021D00005/WH/WHI NIH HHS/United States
- MR/S003061/1/MRC_/Medical Research Council/United Kingdom
- R01 DK114183/DK/NIDDK NIH HHS/United States
- 75N92021D00003/WH/WHI NIH HHS/United States
- R01 HL136574/HL/NHLBI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- MC_UU_00006/1/MRC_/Medical Research Council/United Kingdom
- 75N92021D00004/WH/WHI NIH HHS/United States
- 75N92021D00002/HL/NHLBI NIH HHS/United States
- MR/V027638/1/MRC_/Medical Research Council/United Kingdom
- MR/S004068/2/MRC_/Medical Research Council/United Kingdom
- MR/R026408/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12015/1/MRC_/Medical Research Council/United Kingdom
- 75N92021D00001/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
