Cofactor F420: an expanded view of its distribution, biosynthesis and roles in bacteria and archaea
- PMID: 33851978
- PMCID: PMC8498797
- DOI: 10.1093/femsre/fuab021
Cofactor F420: an expanded view of its distribution, biosynthesis and roles in bacteria and archaea
Abstract
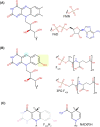
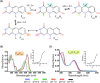
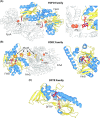
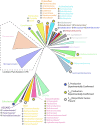
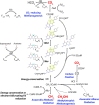
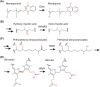
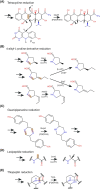
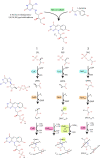
Many bacteria and archaea produce the redox cofactor F420. F420 is structurally similar to the cofactors FAD and FMN but is catalytically more similar to NAD and NADP. These properties allow F420 to catalyze challenging redox reactions, including key steps in methanogenesis, antibiotic biosynthesis and xenobiotic biodegradation. In the last 5 years, there has been much progress in understanding its distribution, biosynthesis, role and applications. Whereas F420 was previously thought to be confined to Actinobacteria and Euryarchaeota, new evidence indicates it is synthesized across the bacterial and archaeal domains, as a result of extensive horizontal and vertical biosynthetic gene transfer. F420 was thought to be synthesized through one biosynthetic pathway; however, recent advances have revealed variants of this pathway and have resolved their key biosynthetic steps. In parallel, new F420-dependent biosynthetic and metabolic processes have been discovered. These advances have enabled the heterologous production of F420 and identified enantioselective F420H2-dependent reductases for biocatalysis. New research has also helped resolve how microorganisms use F420 to influence human and environmental health, providing opportunities for tuberculosis treatment and methane mitigation. A total of 50 years since its discovery, multiple paradigms associated with F420 have shifted, and new F420-dependent organisms and processes continue to be discovered.
Keywords: cofactor 420; cofactor biosynthesis; cofactor distribution; enzymology; redox chemistry; redox cofactor.
© The Author(s) 2021. Published by Oxford University Press on behalf of FEMS.
Figures


References
-
- Abken H-J, Deppenmeier U. Purification and properties of an F420H2 dehydrogenase from Methanosarcina mazei Gö1. FEMS Microbiol Lett. 1997;154:231–7.
-
- Adam PS, Borrel G, Gribaldo S. An archaeal origin of the Wood–Ljungdahl H4MPT branch and the emergence of bacterial methylotrophy. Nat Microbiol. 2019;4:2155–63. - PubMed
-
- Afting C, Hochheimer A, Thauer R. Function of H2-forming methylenetetrahydromethanopterin dehydrogenase from Methanobacterium thermoautotrophicum in coenzyme F420 reduction with H2. Arch Microbiol. 1998;169:206–10. - PubMed
-
- Ahmed FH, Carr PD, Lee BMet al. Sequence–structure–function classification of a catalytically diverse oxidoreductase superfamily in Mycobacteria. J Mol Biol. 2015;427:3554–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

