High-dimensional profiling clusters asthma severity by lymphoid and non-lymphoid status
- PMID: 33852838
- PMCID: PMC8133874
- DOI: 10.1016/j.celrep.2021.108974
High-dimensional profiling clusters asthma severity by lymphoid and non-lymphoid status
Abstract
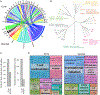
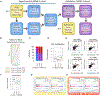
Clinical definitions of asthma fail to capture the heterogeneity of immune dysfunction in severe, treatment-refractory disease. Applying mass cytometry and machine learning to bronchoalveolar lavage (BAL) cells, we find that corticosteroid-resistant asthma patients cluster largely into two groups: one enriched in interleukin (IL)-4+ innate immune cells and another dominated by interferon (IFN)-γ+ T cells, including tissue-resident memory cells. In contrast, BAL cells of a healthier population are enriched in IL-10+ macrophages. To better understand cellular mediators of severe asthma, we developed the Immune Cell Linkage through Exploratory Matrices (ICLite) algorithm to perform deconvolution of bulk RNA sequencing of mixed-cell populations. Signatures of mitosis and IL-7 signaling in CD206-FcεRI+CD127+IL-4+ innate cells in one patient group, contrasting with adaptive immune response in T cells in the other, are preserved across technologies. Transcriptional signatures uncovered by ICLite identify T-cell-high and T-cell-poor severe asthma patients in an independent cohort, suggesting broad applicability of our findings.
Keywords: BAL; CyTOF; FceRI+; ICLite; IFN-g; RNA-seq; clusters; immune; multi-omics; severe asthma.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.R. has a research agreement with Pieris Pharmaceuticals. S.E.W. is a consultant for AstraZeneca, Glaxo Smith-Kline, and Sanofi. She is also involved in clinical trials being run by Knopp, Sanofi, and AstraZeneca. She has a research agreement with Pieris Pharmaceuticals. M.J.C. is a consultant for Pieris Pharmaceuticals.
Figures





References
-
- Alexa A, and Rahnenfuhrer J (2020). topGO: Enrichment analysis for gene ontology (Bioconductor). https://bioconductor.org/packages/release/bioc/html/topGO.html.
-
- Ariotti S, Hogenbirk MA, Dijkgraaf FE, Visser LL, Hoekstra ME, Song JY, Jacobs H, Haanen JB, and Schumacher TN (2014). T cell memory. Skin-resident memory CD8+ T cells trigger a state of tissue-wide pathogen alert. Science 346, 101–105. - PubMed
-
- Aversa G, Chang CC, Carballido JM, Cocks BG, and de Vries JE (1997). Engagement of the signaling lymphocytic activation molecule (SLAM) on activated T cells results in IL-2-independent, cyclosporin A-sensitive T cell proliferation and IFN-gamma production. J. Immunol 158, 4036–4044. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

