Single-cell transcriptomics identifies gene expression networks driving differentiation and tumorigenesis in the human fallopian tube
- PMID: 33852846
- PMCID: PMC10108902
- DOI: 10.1016/j.celrep.2021.108978
Single-cell transcriptomics identifies gene expression networks driving differentiation and tumorigenesis in the human fallopian tube
Abstract
The human fallopian tube harbors the cell of origin for the majority of high-grade serous "ovarian" cancers (HGSCs), but its cellular composition, particularly the epithelial component, is poorly characterized. We perform single-cell transcriptomic profiling of around 53,000 individual cells from 12 primary fallopian specimens to map their major cell types. We identify 10 epithelial subpopulations with diverse transcriptional programs. Based on transcriptional signatures, we reconstruct a trajectory whereby secretory cells differentiate into ciliated cells via a RUNX3high intermediate. Computational deconvolution of advanced HGSCs identifies the "early secretory" population as a likely precursor state for the majority of HGSCs. Its signature comprises both epithelial and mesenchymal features and is enriched in mesenchymal-type HGSCs (p = 6.7 × 10-27), a group known to have particularly poor prognoses. This cellular and molecular compendium of the human fallopian tube in cancer-free women is expected to advance our understanding of the earliest stages of fallopian epithelial neoplasia.
Keywords: PAX8; RUNX3; SOX17; ciliated epithelial cells; fallopian tube; microenvironment; ovarian cancer; scRNA-seq; secretory epithelial cells; transcription factor.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

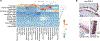




References
-
- Barghout SH, Zepeda N, Vincent K, Azad AK, Xu Z, Yang C, Steed H, Postovit L-M, and Fu Y (2015). RUNX3 contributes to carboplatin resistance in epithelial ovarian cancer cells. Gynecol. Oncol 138, 647–655. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

