Functional analyses of human LUC7-like proteins involved in splicing regulation and myeloid neoplasms
- PMID: 33852859
- PMCID: PMC8078730
- DOI: 10.1016/j.celrep.2021.108989
Functional analyses of human LUC7-like proteins involved in splicing regulation and myeloid neoplasms
Abstract
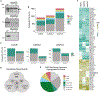
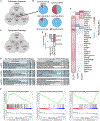
Vertebrates have evolved three paralogs, termed LUC7L, LUC7L2, and LUC7L3, of the essential yeast U1 small nuclear RNA (snRNA)-associated splicing factor Luc7p. We investigated the mechanistic and regulatory functions of these putative splicing factors, of which one (LUC7L2) is mutated or deleted in myeloid neoplasms. Protein interaction data show that all three proteins bind similar core but distinct regulatory splicing factors, probably mediated through their divergent arginine-serine-rich domains, which are not present in Luc7p. Knockdown of each factor reveals mostly unique sets of significantly dysregulated alternative splicing events dependent on their binding locations, which are largely non-overlapping. Notably, knockdown of LUC7L2 alone significantly upregulates the expression of multiple spliceosomal factors and downregulates glycolysis genes, possibly contributing to disease pathogenesis. RNA binding studies reveal that LUC7L2 and LUC7L3 crosslink to weak 5' splice sites and to the 5' end of U1 snRNA, establishing an evolutionarily conserved role in 5' splice site selection.
Keywords: 5′ splice site; LUC7L; LUC7L2; LUC7L3; alternative splicing; myeloid neoplasms.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Comment in
-
When cells are down on their LUC7L2, alternative splicing rewires metabolism for OXPHOS.Mol Cell. 2021 May 6;81(9):1859-1860. doi: 10.1016/j.molcel.2021.04.003. Mol Cell. 2021. PMID: 33961773
References
-
- Bertram K, Agafonov DE, Dybkov O, Haselbach D, Leelaram MN, Will CL, Urlaub H, Kastner B, Lührmann R, and Stark H (2017). Cryo-EM Structure of a Pre-catalytic Human Spliceosome Primed for Activation. Cell 170, 701–713. e11. - PubMed
-
- Bushnell B (2014). BBMap: a fast, accurate, splice-aware aligner (Ernest Orlando Lawrence Berkeley National Laboratory).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

