PRMT1-mediated H4R3me2a recruits SMARCA4 to promote colorectal cancer progression by enhancing EGFR signaling
- PMID: 33853662
- PMCID: PMC8048298
- DOI: 10.1186/s13073-021-00871-5
PRMT1-mediated H4R3me2a recruits SMARCA4 to promote colorectal cancer progression by enhancing EGFR signaling
Erratum in
-
Correction to: PRMT1-mediated H4R3me2a recruits SMARCA4 to promote colorectal cancer progression by enhancing EGFR signaling.Genome Med. 2021 Oct 4;13(1):154. doi: 10.1186/s13073-021-00966-z. Genome Med. 2021. PMID: 34607606 Free PMC article. No abstract available.
Abstract
Background: Aberrant changes in epigenetic mechanisms such as histone modifications play an important role in cancer progression. PRMT1 which triggers asymmetric dimethylation of histone H4 on arginine 3 (H4R3me2a) is upregulated in human colorectal cancer (CRC) and is essential for cell proliferation. However, how this dysregulated modification might contribute to malignant transitions of CRC remains poorly understood.
Methods: In this study, we integrated biochemical assays including protein interaction studies and chromatin immunoprecipitation (ChIP), cellular analysis including cell viability, proliferation, colony formation, and migration assays, clinical sample analysis, microarray experiments, and ChIP-Seq data to investigate the potential genomic recognition pattern of H4R3me2s in CRC cells and its effect on CRC progression.
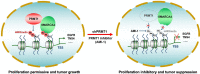
Results: We show that PRMT1 and SMARCA4, an ATPase subunit of the SWI/SNF chromatin remodeling complex, act cooperatively to promote colorectal cancer (CRC) progression. We find that SMARCA4 is a novel effector molecule of PRMT1-mediated H4R3me2a. Mechanistically, we show that H4R3me2a directly recruited SMARCA4 to promote the proliferative, colony-formative, and migratory abilities of CRC cells by enhancing EGFR signaling. We found that EGFR and TNS4 were major direct downstream transcriptional targets of PRMT1 and SMARCA4 in colon cells, and acted in a PRMT1 methyltransferase activity-dependent manner to promote CRC cell proliferation. In vivo, knockdown or inhibition of PRMT1 profoundly attenuated the growth of CRC cells in the C57BL/6 J-ApcMin/+ CRC mice model. Importantly, elevated expression of PRMT1 or SMARCA4 in CRC patients were positively correlated with expression of EGFR and TNS4, and CRC patients had shorter overall survival. These findings reveal a critical interplay between epigenetic and transcriptional control during CRC progression, suggesting that SMARCA4 is a novel key epigenetic modulator of CRC. Our findings thus highlight PRMT1/SMARCA4 inhibition as a potential therapeutic intervention strategy for CRC.
Conclusion: PRMT1-mediated H4R3me2a recruits SMARCA4, which promotes colorectal cancer progression by enhancing EGFR signaling.
Keywords: Colorectal Cancer; Epigenomics; H4R3me2s; PRMT1; SMARCA4; Transcription.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

