Assessment of the microbiome during bacteriophage therapy in combination with systemic antibiotics to treat a case of staphylococcal device infection
- PMID: 33853672
- PMCID: PMC8048313
- DOI: 10.1186/s40168-021-01026-9
Assessment of the microbiome during bacteriophage therapy in combination with systemic antibiotics to treat a case of staphylococcal device infection
Abstract
Background: Infectious bacterial diseases exhibiting increasing resistance to antibiotics are a serious global health issue. Bacteriophage therapy is an anti-microbial alternative to treat patients with serious bacterial infections. However, the impacts to the host microbiome in response to clinical use of phage therapy are not well understood.
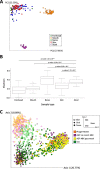
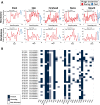
Results: Our paper demonstrates a largely unchanged microbiota profile during 4 weeks of phage therapy when added to systemic antibiotics in a single patient with Staphylococcus aureus device infection. Metabolomic analyses suggest potential indirect cascading ecological impacts to the host (skin) microbiome. We did not detect genomes of the three phages used to treat the patient in metagenomic samples taken from saliva, stool, and skin; however, phages were detected using endpoint-PCR in patient serum.
Conclusion: Results from our proof-of-principal study supports the use of bacteriophages as a microbiome-sparing approach to treat bacterial infections. Video abstract.
Keywords: Bacteriophages; Metabolomics; Microbiome; Phage therapy; Staphylococcus aureus.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures
References
-
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance 2014. http://www.who.int/drugresistance/documents/surveillancereport/en/ Accessed on 22 July 2019
-
- Schooley RT, Biswas B, Gill JJ, et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 2017;61(10):e00954–e00917. doi: 10.1128/AAC.00954-17. - DOI - PMC - PubMed
-
- Aslam S, Lampley E, Wooten D, et al. Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug-resistant bacterial infections at a single center in the United States. Open Forum Infect Dis. 2020;7(9):ofaa389. doi: 10.1093/ofid/ofaa389. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

