Profiling single-cell histone modifications using indexing chromatin immunocleavage sequencing
- PMID: 33853847
- PMCID: PMC8494230
- DOI: 10.1101/gr.260893.120
Profiling single-cell histone modifications using indexing chromatin immunocleavage sequencing
Abstract
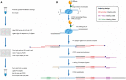
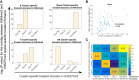
Recently, multiple single-cell assays were developed for detecting histone marks at the single-cell level. These techniques are either limited by the low cell throughput or sparse reads which limit their applications. To address these problems, we introduce indexing single-cell immunocleavage sequencing (iscChIC-seq), a multiplex indexing method based on TdT terminal transferase and T4 DNA ligase-mediated barcoding strategy and single-cell ChIC-seq, which is capable of readily analyzing histone modifications across tens of thousands of single cells in one experiment. Application of iscChIC-seq to profiling H3K4me3 and H3K27me3 in human white blood cells (WBCs) enabled successful detection of more than 10,000 single cells for each histone modification with 11 K and 45 K nonredundant reads per cell, respectively. Cluster analysis of these data allowed identification of monocytes, T cells, B cells, and NK cells from WBCs. The cell types annotated from H3K4me3 single-cell data are specifically correlated with the cell types annotated from H3K27me3 single-cell data. Our data indicate that iscChIC-seq is a reliable technique for profiling histone modifications in a large number of single cells, which may find broad applications in studying cellular heterogeneity and differentiation status in complex developmental and disease systems.
Published by Cold Spring Harbor Laboratory Press.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
