Defining super-enhancer landscape in triple-negative breast cancer by multiomic profiling
- PMID: 33854062
- PMCID: PMC8046763
- DOI: 10.1038/s41467-021-22445-0
Defining super-enhancer landscape in triple-negative breast cancer by multiomic profiling
Abstract
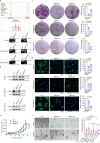
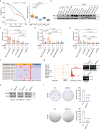
Breast cancer is a heterogeneous disease, affecting over 3.5 million women worldwide, yet the functional role of cis-regulatory elements including super-enhancers in different breast cancer subtypes remains poorly characterized. Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer with a poor prognosis. Here we apply integrated epigenomic and transcriptomic profiling to uncover super-enhancer heterogeneity between breast cancer subtypes, and provide clinically relevant biological insights towards TNBC. Using CRISPR/Cas9-mediated gene editing, we identify genes that are specifically regulated by TNBC-specific super-enhancers, including FOXC1 and MET, thereby unveiling a mechanism for specific overexpression of the key oncogenes in TNBC. We also identify ANLN as a TNBC-specific gene regulated by super-enhancer. Our studies reveal a TNBC-specific epigenomic landscape, contributing to the dysregulated oncogene expression in breast tumorigenesis.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J. Clinicians. 2018;68:394–424. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

