SIRT1 promotes lipid metabolism and mitochondrial biogenesis in adipocytes and coordinates adipogenesis by targeting key enzymatic pathways
- PMID: 33854178
- PMCID: PMC8046990
- DOI: 10.1038/s41598-021-87759-x
SIRT1 promotes lipid metabolism and mitochondrial biogenesis in adipocytes and coordinates adipogenesis by targeting key enzymatic pathways
Abstract
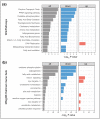
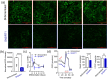
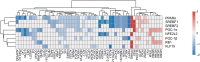
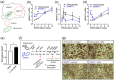
The NAD+-dependent deacetylase SIRT1 controls key metabolic functions by deacetylating target proteins and strategies that promote SIRT1 function such as SIRT1 overexpression or NAD+ boosters alleviate metabolic complications. We previously reported that SIRT1-depletion in 3T3-L1 preadipocytes led to C-Myc activation, adipocyte hyperplasia, and dysregulated adipocyte metabolism. Here, we characterized SIRT1-depleted adipocytes by quantitative mass spectrometry-based proteomics, gene-expression and biochemical analyses, and mitochondrial studies. We found that SIRT1 promoted mitochondrial biogenesis and respiration in adipocytes and expression of molecules like leptin, adiponectin, matrix metalloproteinases, lipocalin 2, and thyroid responsive protein was SIRT1-dependent. Independent validation of the proteomics dataset uncovered SIRT1-dependence of SREBF1c and PPARα signaling in adipocytes. SIRT1 promoted nicotinamide mononucleotide acetyltransferase 2 (NMNAT2) expression during 3T3-L1 differentiation and constitutively repressed NMNAT1 and 3 levels. Supplementing preadipocytes with the NAD+ booster nicotinamide mononucleotide (NMN) during differentiation increased expression levels of leptin, SIRT1, and PGC-1α and its transcriptional targets, and reduced levels of pro-fibrotic collagens (Col6A1 and Col6A3) in a SIRT1-dependent manner. Investigating the metabolic impact of the functional interaction of SIRT1 with SREBF1c and PPARα and insights into how NAD+ metabolism modulates adipocyte function could potentially lead to new avenues in developing therapeutics for obesity complications.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

