CRL4AMBRA1 is a master regulator of D-type cyclins
- PMID: 33854235
- PMCID: PMC8875297
- DOI: 10.1038/s41586-021-03445-y
CRL4AMBRA1 is a master regulator of D-type cyclins
Abstract
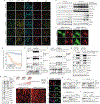
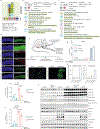
D-type cyclins are central regulators of the cell division cycle and are among the most frequently deregulated therapeutic targets in human cancer1, but the mechanisms that regulate their turnover are still being debated2,3. Here, by combining biochemical and genetics studies in somatic cells, we identify CRL4AMBRA1 (also known as CRL4DCAF3) as the ubiquitin ligase that targets all three D-type cyclins for degradation. During development, loss of Ambra1 induces the accumulation of D-type cyclins and retinoblastoma (RB) hyperphosphorylation and hyperproliferation, and results in defects of the nervous system that are reduced by treating pregnant mice with the FDA-approved CDK4 and CDK6 (CDK4/6) inhibitor abemaciclib. Moreover, AMBRA1 acts as a tumour suppressor in mouse models and low AMBRA1 mRNA levels are predictive of poor survival in cancer patients. Cancer hotspot mutations in D-type cyclins abrogate their binding to AMBRA1 and induce their stabilization. Finally, a whole-genome, CRISPR-Cas9 screen identified AMBRA1 as a regulator of the response to CDK4/6 inhibition. Loss of AMBRA1 reduces sensitivity to CDK4/6 inhibitors by promoting the formation of complexes of D-type cyclins with CDK2. Collectively, our results reveal the molecular mechanism that controls the stability of D-type cyclins during cell-cycle progression, in development and in human cancer, and implicate AMBRA1 as a critical regulator of the RB pathway.
Conflict of interest statement
Figures











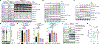


References
-
- Malumbres M & Barbacid M Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153–166 (2009). - PubMed
-
- Reichermeier KM et al. PIKES analysis reveals response to degraders and key regulatory mechanisms of the CRL4 network. Mol. Cell 77, 1092–1106.e9 (2020). - PubMed
-
- Natsume T, Kiyomitsu T, Saga Y & Kanemaki MT Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 15, 210–218 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

