Antibiotics and Antibiotic Resistance Genes in Animal Manure - Consequences of Its Application in Agriculture
- PMID: 33854486
- PMCID: PMC8039466
- DOI: 10.3389/fmicb.2021.610656
Antibiotics and Antibiotic Resistance Genes in Animal Manure - Consequences of Its Application in Agriculture
Abstract
Antibiotic resistance genes (ARGs) are a relatively new type of pollutant. The rise in antibiotic resistance observed recently is closely correlated with the uncontrolled and widespread use of antibiotics in agriculture and the treatment of humans and animals. Resistant bacteria have been identified in soil, animal feces, animal housing (e.g., pens, barns, or pastures), the areas around farms, manure storage facilities, and the guts of farm animals. The selection pressure caused by the irrational use of antibiotics in animal production sectors not only promotes the survival of existing antibiotic-resistant bacteria but also the development of new resistant forms. One of the most critical hot-spots related to the development and dissemination of ARGs is livestock and poultry production. Manure is widely used as a fertilizer thanks to its rich nutrient and organic matter content. However, research indicates that its application may pose a severe threat to human and animal health by facilitating the dissemination of ARGs to arable soil and edible crops. This review examines the pathogens, potentially pathogenic microorganisms and ARGs which may be found in animal manure, and evaluates their effect on human health through their exposure to soil and plant resistomes. It takes a broader view than previous studies of this topic, discussing recent data on antibiotic use in farm animals and the effect of these practices on the composition of animal manure; it also examines how fertilization with animal manure may alter soil and crop microbiomes, and proposes the drivers of such changes and their consequences for human health.
Keywords: animal agriculture; antibiotic resistance genes; antibiotic use; antibiotic-resistant bacteria; fecal matter; manure resistome; plant resistome; soil resistome.
Copyright © 2021 Zalewska, Błażejewska, Czapko and Popowska.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

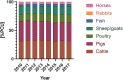
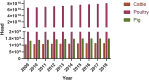
References
-
- Abriouel H., Omar N. B., Molinos A. C., López R. L., Grande M. J., Martínez-Viedma P., et al. (2008). Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. Int. J. Food Microbiol. 123 38–49. 10.1016/j.ijfoodmicro.2007.11.067 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

