Streptococcal protein SIC activates monocytes and induces inflammation
- PMID: 33855284
- PMCID: PMC8027542
- DOI: 10.1016/j.isci.2021.102339
Streptococcal protein SIC activates monocytes and induces inflammation
Abstract
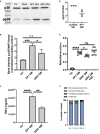
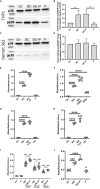
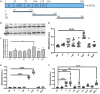
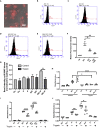
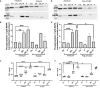
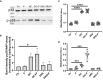
Streptococcus pyogenes is a major bacterial pathogen in the human population and isolates of the clinically important M1 serotype secrete protein Streptococcal inhibitor of complement (SIC) known to interfere with human innate immunity. Here we find that SIC from M1 bacteria interacts with TLR2 and CD14 on monocytes leading to the activation of the NF-κB and p38 MAPK pathways and the release of several pro-inflammatory cytokines (e.g. TNFα and INFγ). In human plasma, SIC binds clusterin and histidine-rich glycoprotein, and whole plasma, and these two purified plasma proteins enhanced the activation of monocytes by SIC. Isolates of the M55 serotype secrete an SIC homolog, but this protein did not activate monocytes. M1 isolates are common in cases of invasive S. pyogenes infections characterized by massive inflammation, and the results of this study indicate that the pro-inflammatory property of SIC contributes to the pathology of these severe clinical conditions.
Keywords: Clinical Microbiology; Immunology; Microbiology.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
Streptococcal inhibitor of complement (SIC) modulates fibrinolysis and enhances bacterial survival within fibrin clots.J Biol Chem. 2018 Aug 31;293(35):13578-13591. doi: 10.1074/jbc.RA118.001988. Epub 2018 Jul 12. J Biol Chem. 2018. PMID: 30002122 Free PMC article.
-
Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function.J Biol Chem. 1996 Jan 12;271(2):1081-8. doi: 10.1074/jbc.271.2.1081. J Biol Chem. 1996. PMID: 8557634
-
Streptococcal inhibitor of complement-mediated lysis (SIC): an anti-inflammatory virulence determinant.Microbiology (Reading). 2010 Dec;156(Pt 12):3660-3668. doi: 10.1099/mic.0.039578-0. Epub 2010 Aug 12. Microbiology (Reading). 2010. PMID: 20705662 Free PMC article.
-
Antibacterial activity of the contact and complement systems is blocked by SIC, a protein secreted by Streptococcus pyogenes.J Biol Chem. 2011 Jan 14;286(2):1331-40. doi: 10.1074/jbc.M110.178350. Epub 2010 Nov 10. J Biol Chem. 2011. PMID: 21068386 Free PMC article.
-
Vibrio harveyi infections induce production of proinflammatory cytokines in murine peritoneal macrophages via activation of p38 MAPK and NF-κB pathways, but reversed by PI3K/AKT pathways.Dev Comp Immunol. 2022 Feb;127:104292. doi: 10.1016/j.dci.2021.104292. Epub 2021 Oct 14. Dev Comp Immunol. 2022. PMID: 34656643 Review.
Cited by
-
Immune signature of acute pharyngitis in a Streptococcus pyogenes human challenge trial.Nat Commun. 2022 Feb 9;13(1):769. doi: 10.1038/s41467-022-28335-3. Nat Commun. 2022. PMID: 35140232 Free PMC article.
-
Additional biomarkers and emm types associated with group A streptococcal toxic shock syndrome: a Japanese nationwide observational study.Eur J Clin Microbiol Infect Dis. 2025 Jun;44(6):1471-1480. doi: 10.1007/s10096-025-05116-6. Epub 2025 Apr 7. Eur J Clin Microbiol Infect Dis. 2025. PMID: 40193020 Free PMC article.
-
Analysis of host-pathogen gene association networks reveals patient-specific response to streptococcal and polymicrobial necrotising soft tissue infections.BMC Med. 2022 May 4;20(1):173. doi: 10.1186/s12916-022-02355-8. BMC Med. 2022. PMID: 35505341 Free PMC article.
-
Complement evasion factor (CEF), a novel immune evasion factor of Streptococcus pyogenes.Virulence. 2022 Dec;13(1):225-240. doi: 10.1080/21505594.2022.2027629. Virulence. 2022. PMID: 35094646 Free PMC article.
-
Modulatory Effects of the Recombinant Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Spike S1 Subunit Protein on the Phenotype of Camel Monocyte-Derived Macrophages.Biology (Basel). 2025 Mar 13;14(3):292. doi: 10.3390/biology14030292. Biology (Basel). 2025. PMID: 40136548 Free PMC article.
References
-
- Åkesson P., Herwald H., Rasmussen M., HÅkansson K., Abrahamson M., Hasan A.A.K., Schmaier A.H., Müller-Esterl W., Björck L. Streptococcal inhibitor of complement-mediated lysis (SIC): an anti-inflammatory virulence determinant. Microbiology. 2010;156:3660–3668. doi: 10.1099/mic.0.039578-0. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

