Time-resolved proteomics profiling of the ciliary Hedgehog response
- PMID: 33856408
- PMCID: PMC8054476
- DOI: 10.1083/jcb.202007207
Time-resolved proteomics profiling of the ciliary Hedgehog response
Abstract
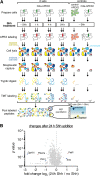
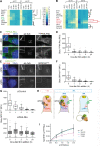
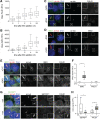
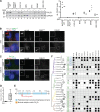
The primary cilium is a signaling compartment that interprets Hedgehog signals through changes of its protein, lipid, and second messenger compositions. Here, we combine proximity labeling of cilia with quantitative mass spectrometry to unbiasedly profile the time-dependent alterations of the ciliary proteome in response to Hedgehog. This approach correctly identifies the three factors known to undergo Hedgehog-regulated ciliary redistribution and reveals two such additional proteins. First, we find that a regulatory subunit of the cAMP-dependent protein kinase (PKA) rapidly exits cilia together with the G protein-coupled receptor GPR161 in response to Hedgehog, and we propose that the GPR161/PKA module senses and amplifies cAMP signals to modulate ciliary PKA activity. Second, we identify the phosphatase Paladin as a cell type-specific regulator of Hedgehog signaling that enters primary cilia upon pathway activation. The broad applicability of quantitative ciliary proteome profiling promises a rapid characterization of ciliopathies and their underlying signaling malfunctions.
© 2021 May et al.
Figures







References
-
- Amieux, P.S., Howe D.G., Knickerbocker H., Lee D.C., Su T., Laszlo G.S., Idzerda R.L., and McKnight G.S.. 2002. Increased basal cAMP-dependent protein kinase activity inhibits the formation of mesoderm-derived structures in the developing mouse embryo. J. Biol. Chem. 277:27294–27304. 10.1074/jbc.M200302200 - DOI - PubMed
-
- Arveseth, C.D., Happ J.T., Hedeen D.S., Zhu J.-F., Capener J.L., Shaw D.K., Deshpande I., Liang J., Xu J., Stubben S.L., et al. . 2020. Smoothened Transduces Hedgehog Signals via Activity-Dependent Sequestration of PKA Catalytic Subunits. bioRxiv. doi: 10.1101/2020.07.01.183079 (Preprint posted July 2, 2020). - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

