Structure of the E. coli agmatinase, SPEB
- PMID: 33857156
- PMCID: PMC8049259
- DOI: 10.1371/journal.pone.0248991
Structure of the E. coli agmatinase, SPEB
Abstract
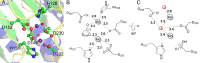
Agmatine amidinohydrolase, or agmatinase, catalyzes the conversion of agmatine to putrescine and urea. This enzyme is found broadly across kingdoms of life and plays a critical role in polyamine biosynthesis and the regulation of agmatine concentrations. Here we describe the high-resolution X-ray crystal structure of the E. coli agmatinase, SPEB. The data showed a relatively high degree of pseudomerohedral twinning, was ultimately indexed in the P31 space group and led to a final model with eighteen chains, corresponding to three full hexamers in the asymmetric unit. There was a solvent content of 38.5% and refined R/Rfree values of 0.166/0.216. The protein has the conserved fold characteristic of the agmatine ureohydrolase family and displayed a high degree of structural similarity among individual protomers. Two distinct peaks of electron density were observed in the active site of most of the eighteen chains of SPEB. As the activity of this protein is known to be dependent upon manganese and the fold is similar to other dinuclear metallohydrolases, these peaks were modeled as manganese ions. The orientation of the conserved active site residues, in particular those amino acids that participate in binding the metal ions and a pair of acidic residues (D153 and E274 in SPEB) that play a role in catalysis, are similar to other agmatinase and arginase enzymes and is consistent with a hydrolytic mechanism that proceeds via a metal-activated hydroxide ion.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

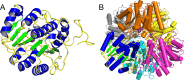


Similar articles
-
Crystal Structure of Escherichia coli Agmatinase: Catalytic Mechanism and Residues Relevant for Substrate Specificity.Int J Mol Sci. 2021 Apr 30;22(9):4769. doi: 10.3390/ijms22094769. Int J Mol Sci. 2021. PMID: 33946272 Free PMC article.
-
Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily.J Biol Chem. 2004 Nov 26;279(48):50505-13. doi: 10.1074/jbc.M409246200. Epub 2004 Sep 7. J Biol Chem. 2004. PMID: 15355972
-
Insights into the Mn2+ Binding Site in the Agmatinase-Like Protein (ALP): A Critical Enzyme for the Regulation of Agmatine Levels in Mammals.Int J Mol Sci. 2020 Jun 10;21(11):4132. doi: 10.3390/ijms21114132. Int J Mol Sci. 2020. PMID: 32531922 Free PMC article.
-
Functional analysis of the Mn2+ requirement in the catalysis of ureohydrolases arginase and agmatinase - a historical perspective.J Inorg Biochem. 2020 Jan;202:110812. doi: 10.1016/j.jinorgbio.2019.110812. Epub 2019 Aug 26. J Inorg Biochem. 2020. PMID: 31731096 Review.
-
Vertebrate agmatinases: what role do they play in agmatine catabolism?Ann N Y Acad Sci. 2003 Dec;1009:30-3. doi: 10.1196/annals.1304.003. Ann N Y Acad Sci. 2003. PMID: 15028567 Review.
Cited by
-
Large-scale phylogenomics of aquatic bacteria reveal molecular mechanisms for adaptation to salinity.Sci Adv. 2023 May 26;9(21):eadg2059. doi: 10.1126/sciadv.adg2059. Epub 2023 May 26. Sci Adv. 2023. PMID: 37235649 Free PMC article.
-
The Pseudomonas aeruginosa Type VI secretion system toxin Tse8 evolved from a novel N-carbamoylputrescine amidohydrolase.Biochem J. 2025 Jul 22;482(15):BCJ20253210. doi: 10.1042/BCJ20253210. Biochem J. 2025. PMID: 40673658 Free PMC article.
-
Metabolic potential of Nitrososphaera-associated clades.ISME J. 2024 Jan 8;18(1):wrae086. doi: 10.1093/ismejo/wrae086. ISME J. 2024. PMID: 38742714 Free PMC article.
-
Gut microbiota bridges dietary nutrients and host immunity.Sci China Life Sci. 2023 Nov;66(11):2466-2514. doi: 10.1007/s11427-023-2346-1. Epub 2023 Jun 5. Sci China Life Sci. 2023. PMID: 37286860 Free PMC article.
-
MetaBiome: a multiscale model integrating agent-based and metabolic networks to reveal spatial regulation in gut mucosal microbial communities.mSystems. 2025 May 20;10(5):e0165224. doi: 10.1128/msystems.01652-24. Epub 2025 Apr 4. mSystems. 2025. PMID: 40183581 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

