N- and C-terminal Gln3-Tor1 interaction sites: one acting negatively and the other positively to regulate nuclear Gln3 localization
- PMID: 33857304
- PMCID: PMC8049557
- DOI: 10.1093/genetics/iyab017
N- and C-terminal Gln3-Tor1 interaction sites: one acting negatively and the other positively to regulate nuclear Gln3 localization
Abstract
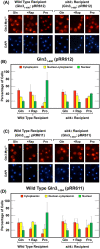
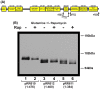
Gln3 activates Nitrogen Catabolite Repression, NCR-sensitive expression of the genes required for Saccharomyces cerevisiae to scavenge poor nitrogen sources from its environment. The global TorC1 kinase complex negatively regulates nuclear Gln3 localization, interacting with an α-helix in the C-terminal region of Gln3, Gln3656-666. In nitrogen replete conditions, Gln3 is sequestered in the cytoplasm, whereas when TorC1 is down-regulated, in nitrogen restrictive conditions, Gln3 migrates into the nucleus. In this work, we show that the C-terminal Gln3-Tor1 interaction site is required for wild type, rapamycin-elicited, Sit4-dependent nuclear Gln3 localization, but not for its dephosphorylation. In fact, truncated Gln31-384 can enter the nucleus in the absence of Sit4 in both repressive and derepressive growth conditions. However, Gln31-384 can only enter the nucleus if a newly discovered second positively-acting Gln3-Tor1 interaction site remains intact. Importantly, the N- and C-terminal Gln3-Tor1 interaction sites function both autonomously and collaboratively. The N-terminal Gln3-Tor1 interaction site, previously designated Gln3URS contains a predicted α-helix situated within an unstructured coiled-coil region. Eight of the thirteen serine/threonine residues in the Gln3URS are dephosphorylated 3-15-fold with three of them by 10-15-fold. Substituting phosphomimetic aspartate for serine/threonine residues in the Gln3 URS abolishes the N-terminal Gln3-Tor1 interaction, rapamycin-elicited nuclear Gln3 localization, and ½ of the derepressed levels of nuclear Gln3 localization. Cytoplasmic Gln3 sequestration in repressive conditions, however, remains intact. These findings further deconvolve the mechanisms that achieve nitrogen-responsive transcription factor regulation downstream of TorC1.
Keywords: Gln3; Nitrogen metabolism; Signal transduction; Sit4; TOR complex (TorC1); Transcription factors; URS.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures











Similar articles
-
Sit4 and PP2A Dephosphorylate Nitrogen Catabolite Repression-Sensitive Gln3 When TorC1 Is Up- as Well as Downregulated.Genetics. 2019 Aug;212(4):1205-1225. doi: 10.1534/genetics.119.302371. Epub 2019 Jun 18. Genetics. 2019. PMID: 31213504 Free PMC article.
-
Tor pathway control of the nitrogen-responsive DAL5 gene bifurcates at the level of Gln3 and Gat1 regulation in Saccharomyces cerevisiae.J Biol Chem. 2008 Apr 4;283(14):8919-29. doi: 10.1074/jbc.M708811200. Epub 2008 Feb 1. J Biol Chem. 2008. PMID: 18245087 Free PMC article.
-
A domain in the transcription activator Gln3 specifically required for rapamycin responsiveness.J Biol Chem. 2014 Jul 4;289(27):18999-9018. doi: 10.1074/jbc.M114.563668. Epub 2014 May 20. J Biol Chem. 2014. PMID: 24847055 Free PMC article.
-
Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots.FEMS Microbiol Rev. 2002 Aug;26(3):223-38. doi: 10.1111/j.1574-6976.2002.tb00612.x. FEMS Microbiol Rev. 2002. PMID: 12165425 Free PMC article. Review.
-
The intersection between stress responses and inositol pyrophosphates in Saccharomyces cerevisiae.Curr Genet. 2020 Oct;66(5):901-910. doi: 10.1007/s00294-020-01078-8. Epub 2020 Apr 23. Curr Genet. 2020. PMID: 32322930 Review.
Cited by
-
TORC1 Signaling Controls the Stability and Function of α-Arrestins Aly1 and Aly2.Biomolecules. 2022 Mar 31;12(4):533. doi: 10.3390/biom12040533. Biomolecules. 2022. PMID: 35454122 Free PMC article.
-
TOR Complex 1: Orchestrating Nutrient Signaling and Cell Cycle Progression.Int J Mol Sci. 2023 Oct 30;24(21):15745. doi: 10.3390/ijms242115745. Int J Mol Sci. 2023. PMID: 37958727 Free PMC article. Review.
-
CmNDB1 and a Specific Domain of CmMYB1 Negatively Regulate CmMYB1-Dependent Transcription of Nitrate Assimilation Genes Under Nitrogen-Repleted Condition in a Unicellular Red Alga.Front Plant Sci. 2022 Mar 11;13:821947. doi: 10.3389/fpls.2022.821947. eCollection 2022. Front Plant Sci. 2022. PMID: 35360310 Free PMC article.
-
Disordered sequences of transcription factors regulate genomic binding by integrating diverse sequence grammars and interaction types.Nucleic Acids Res. 2024 Aug 27;52(15):8763-8777. doi: 10.1093/nar/gkae521. Nucleic Acids Res. 2024. PMID: 38908024 Free PMC article.
-
Effects of abolishing Whi2 on the proteome and nitrogen catabolite repression-sensitive protein production.G3 (Bethesda). 2022 Mar 4;12(3):jkab432. doi: 10.1093/g3journal/jkab432. G3 (Bethesda). 2022. PMID: 35100365 Free PMC article.
References
-
- Beck T, Hall MN.. 1999. The TOR signaling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 402:689–692. - PubMed
-
- Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, et al.2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J Biol Chem. 275:35727–35733. - PubMed
-
- Binda M, Bonfils G, Panchaud N, Péli-Gulli MP, De Virgilio C.. 2010. An EGOcentric view of TORC1 signaling. Cell Cycle. 9:221–222. - PubMed
-
- Binda M, Péli-Gulli MP, Bonfils G, Panchaud N, Urban J, et al.2009. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 35:563–573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

