The DHX36-specific-motif (DSM) enhances specificity by accelerating recruitment of DNA G-quadruplex structures
- PMID: 33857359
- PMCID: PMC8054968
- DOI: 10.1515/hsz-2020-0302
The DHX36-specific-motif (DSM) enhances specificity by accelerating recruitment of DNA G-quadruplex structures
Abstract
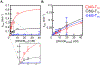
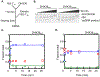
DHX36 is a eukaryotic DEAH/RHA family helicase that disrupts G-quadruplex structures (G4s) with high specificity, contributing to regulatory roles of G4s. Here we used a DHX36 truncation to examine the roles of the 13-amino acid DHX36-specific motif (DSM) in DNA G4 recognition and disruption. We found that the DSM promotes G4 recognition and specificity by increasing the G4 binding rate of DHX36 without affecting the dissociation rate. Further, for most of the G4s measured, the DSM has little or no effect on the G4 disruption step by DHX36, implying that contacts with the G4 are maintained through the transition state for G4 disruption. This result suggests that partial disruption of the G4 from the 3' end is sufficient to reach the overall transition state for G4 disruption, while the DSM remains unperturbed at the 5' end. Interestingly, the DSM does not contribute to G4 binding kinetics or thermodynamics at low temperature, indicating a highly modular function. Together, our results animate recent DHX36 crystal structures, suggesting a model in which the DSM recruits G4s in a modular and flexible manner by contacting the 5' face early in binding, prior to rate-limiting capture and disruption of the G4 by the helicase core.
Keywords: G-quadruplex; G4 resolvase; G4R1; RHAU; RNA helicase; helicase kinetics.
© 2020 Bruce Chang-Gu et al., published by De Gruyter, Berlin/Boston.
Figures
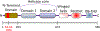





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
