Multi-omic profiling of pituitary thyrotropic cells and progenitors
- PMID: 33858413
- PMCID: PMC8051135
- DOI: 10.1186/s12915-021-01009-0
Multi-omic profiling of pituitary thyrotropic cells and progenitors
Abstract
Background: The pituitary gland is a neuroendocrine organ containing diverse cell types specialized in secreting hormones that regulate physiology. Pituitary thyrotropes produce thyroid-stimulating hormone (TSH), a critical factor for growth and maintenance of metabolism. The transcription factors POU1F1 and GATA2 have been implicated in thyrotrope fate, but the transcriptomic and epigenomic landscapes of these neuroendocrine cells have not been characterized. The goal of this work was to discover transcriptional regulatory elements that drive thyrotrope fate.
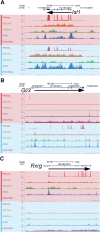
Results: We identified the transcription factors and epigenomic changes in chromatin that are associated with differentiation of POU1F1-expressing progenitors into thyrotropes using cell lines that represent an undifferentiated Pou1f1 lineage progenitor (GHF-T1) and a committed thyrotrope line that produces TSH (TαT1). We compared RNA-seq, ATAC-seq, histone modification (H3K27Ac, H3K4Me1, and H3K27Me3), and POU1F1 binding in these cell lines. POU1F1 binding sites are commonly associated with bZIP transcription factor consensus binding sites in GHF-T1 cells and Helix-Turn-Helix (HTH) or basic Helix-Loop-Helix (bHLH) factors in TαT1 cells, suggesting that these classes of transcription factors may recruit or cooperate with POU1F1 binding at unique sites. We validated enhancer function of novel elements we mapped near Cga, Pitx1, Gata2, and Tshb by transfection in TαT1 cells. Finally, we confirmed that an enhancer element near Tshb can drive expression in thyrotropes of transgenic mice, and we demonstrate that GATA2 enhances Tshb expression through this element.
Conclusion: These results extend the ENCODE multi-omic profiling approach to the pituitary gland, which should be valuable for understanding pituitary development and disease pathogenesis.
Keywords: Chromatin; GATA2; POU1F1; TSH; bHLH; bHTH; bZIP.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Jarvis JP, Scheinfeldt LB, Soi S, Lambert C, Omberg L, Ferwerda B, Froment A, Bodo JM, Beggs W, Hoffman G, Mezey J, Tishkoff SA. Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African pygmies. PLoS Genet. 2012;8(4):e1002641. doi: 10.1371/journal.pgen.1002641. - DOI - PMC - PubMed
-
- Ye Z, Li Z, Wang Y, Mao Y, Shen M, Zhang Q, Li S, Zhou L, Shou X, Chen J, Song Z, Ma Z, Zhang Z, Li Y, Ye H, Huang C, Wang T, He W, Zhang Y, Xie R, Qiao N, Qiu H, Huang S, Wang M, Shen J, Wen Z, Li W, Liu K, Zhou J, Wang L, Ji J, Wang Y, Chen H, Cheng H, Shi Z, Zhu Y, Geng D, Yao Z, Tang W, Lu B, Pan L, Zhang Y, Bao W, Wu J, Zheng K, Shi Y, Zhao Y. Common variants at 10p12.31, 10q21.1 and 13q12.13 are associated with sporadic pituitary adenoma. Nat Genet. 2015;47(7):793–797. doi: 10.1038/ng.3322. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

