The biological function and clinical significance of STIL in osteosarcoma
- PMID: 33858425
- PMCID: PMC8051131
- DOI: 10.1186/s12935-021-01922-y
The biological function and clinical significance of STIL in osteosarcoma
Abstract
Background: SCL/TAL1 interrupting locus (STIL) is associated with the progression of several tumors; however, the biological role of STIL in osteosarcoma remains poorly understood.
Methods: In this study, the clinical significance of STIL in osteosarcoma was analyzed by gene chip data recorded in public databases. STIL expression was silenced in osteosarcoma cell lines to observe the effects on proliferation, apoptosis, invasion, and migration. Differentially expressed genes (DEGs) in the osteosarcoma chip were analyzed using The Limma package, and STIL co-expressed genes were obtained via the Pearson correlation coefficient. The potential molecular mechanism of STIL in osteosarcoma was further explored by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
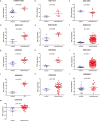
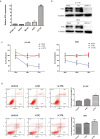
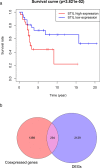
Results: Osteosarcoma was associated with higher STIL expression compared to the control samples, and the standardized mean difference (SMD) was 1.52. STIL also had a good ability to distinguish osteosarcoma from non-osteosarcoma samples [area under the curve (AUC) = 0.96]. After silencing STIL, osteosarcoma cell proliferation decreased, apoptosis increased, and the migratory and invasion ability decreased. A total of 294 STIL differentially co-expressed genes were screened, and a bioinformatics analysis found that differentially co-expressed genes were primarily enriched in the cell signaling pathways. The protein-protein interaction (PPI) network indicated that the hub differentially co-expressed genes of STIL were CDK1, CCNB2, CDC20, CCNA2, BUB1, and AURKB.
Conclusions: STIL is associated with osteosarcoma proliferation and invasion, and may be promote the progression of osteosarcoma by regulating the expression of CDK1, CCNB2, CDC20, CCNA2, BUB1 and AURKB.
Keywords: Biomarkers; Invasion; Osteosarcoma; Proliferation; STIL.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






Similar articles
-
Identification of Key Pathways and Genes in Anaplastic Thyroid Carcinoma via Integrated Bioinformatics Analysis.Med Sci Monit. 2018 Sep 14;24:6438-6448. doi: 10.12659/MSM.910088. Med Sci Monit. 2018. PMID: 30213925 Free PMC article.
-
Identification of key genes associated with bladder cancer using gene expression profiles.Oncol Lett. 2018 Jan;15(1):297-303. doi: 10.3892/ol.2017.7310. Epub 2017 Oct 31. Oncol Lett. 2018. PMID: 29375713 Free PMC article.
-
Examining the biomarkers and molecular mechanisms of medulloblastoma based on bioinformatics analysis.Oncol Lett. 2019 Jul;18(1):433-441. doi: 10.3892/ol.2019.10314. Epub 2019 May 3. Oncol Lett. 2019. PMID: 31289514 Free PMC article.
-
Bioinformatical Analysis of Gene Expression Omnibus Database Associates TAF7/CCNB1, TAF7/CCNA2, and GTF2E2/CDC20 Pathways with Glioblastoma Development and Prognosis.World Neurosurg. 2020 Jun;138:e492-e514. doi: 10.1016/j.wneu.2020.02.159. Epub 2020 Mar 5. World Neurosurg. 2020. PMID: 32147549
-
PODN is a prognostic biomarker and correlated with immune infiltrates in osteosarcoma.Cancer Cell Int. 2021 Jul 17;21(1):381. doi: 10.1186/s12935-021-02086-5. Cancer Cell Int. 2021. PMID: 34273970 Free PMC article. Review.
Cited by
-
O-GlcNAcylation of NONO regulates paraspeckle component assembly and contributes to colon cancer cell proliferation.Cell Death Discov. 2025 May 13;11(1):234. doi: 10.1038/s41420-025-02405-z. Cell Death Discov. 2025. PMID: 40360465 Free PMC article.
-
Lung Cancer Gene Regulatory Network of Transcription Factors Related to the Hallmarks of Cancer.Curr Issues Mol Biol. 2023 Jan 5;45(1):434-464. doi: 10.3390/cimb45010029. Curr Issues Mol Biol. 2023. PMID: 36661515 Free PMC article.
-
Telomeres and telomerase in Sarcoma disease and therapy.Int J Med Sci. 2024 Aug 6;21(11):2065-2080. doi: 10.7150/ijms.97485. eCollection 2024. Int J Med Sci. 2024. PMID: 39239547 Free PMC article. Review.
-
Regulatory factor X-5/SCL/TAL1 interruption site axis promotes aerobic glycolysis and hepatocellular carcinoma cell stemness.Kaohsiung J Med Sci. 2025 Jan;41(1):e12922. doi: 10.1002/kjm2.12922. Epub 2024 Dec 24. Kaohsiung J Med Sci. 2025. PMID: 39718123 Free PMC article.
-
Interaction of STIL with FOXM1 regulates SF3A3 transcription in the hepatocellular carcinoma development.Cell Div. 2025 Jan 17;20(1):1. doi: 10.1186/s13008-025-00142-4. Cell Div. 2025. PMID: 39825314 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

