Democratising deep learning for microscopy with ZeroCostDL4Mic
- PMID: 33859193
- PMCID: PMC8050272
- DOI: 10.1038/s41467-021-22518-0
Democratising deep learning for microscopy with ZeroCostDL4Mic
Abstract
Deep Learning (DL) methods are powerful analytical tools for microscopy and can outperform conventional image processing pipelines. Despite the enthusiasm and innovations fuelled by DL technology, the need to access powerful and compatible resources to train DL networks leads to an accessibility barrier that novice users often find difficult to overcome. Here, we present ZeroCostDL4Mic, an entry-level platform simplifying DL access by leveraging the free, cloud-based computational resources of Google Colab. ZeroCostDL4Mic allows researchers with no coding expertise to train and apply key DL networks to perform tasks including segmentation (using U-Net and StarDist), object detection (using YOLOv2), denoising (using CARE and Noise2Void), super-resolution microscopy (using Deep-STORM), and image-to-image translation (using Label-free prediction - fnet, pix2pix and CycleGAN). Importantly, we provide suitable quantitative tools for each network to evaluate model performance, allowing model optimisation. We demonstrate the application of the platform to study multiple biological processes.
Conflict of interest statement
We provide a platform based on Google Drive and Google Colab to streamline the implementation of common Deep Learning analysis of microscopy data. Despite heavily relying on Google products, we have no commercial or financial interest in promoting and using them. In particular, we did not receive any compensation in any form from Google for this work. The authors declare no competing interests.
Figures

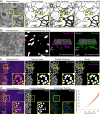

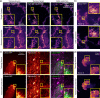

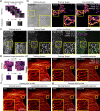
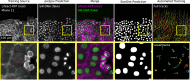


References
-
- Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. in Advances in Neural Information Processing Systems 25 (eds. Pereira, F. et al.) 1097–1105 (Curran Associates, Inc., 2012).
-
- Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention. pp. 234–241 (Springer, Cham, 2015).
-
- Redmon, J. & Farhadi, A. YOLO9000: better, faster, stronger. In Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 7263–7271 (2017).
-
- Schmidt, U., Weigert, M., Broaddus, C. & Myers, G. Cell detection with Star-Convex polygons. in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018 (eds. Frangi, A. F. et al.) Vol. 11071, 265–273 (Springer International Publishing, 2018).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

