Microglial neuropilin-1 promotes oligodendrocyte expansion during development and remyelination by trans-activating platelet-derived growth factor receptor
- PMID: 33859199
- PMCID: PMC8050320
- DOI: 10.1038/s41467-021-22532-2
Microglial neuropilin-1 promotes oligodendrocyte expansion during development and remyelination by trans-activating platelet-derived growth factor receptor
Abstract
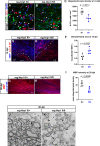
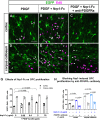
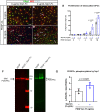
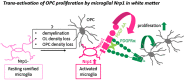
Nerve-glia (NG2) glia or oligodendrocyte precursor cells (OPCs) are distributed throughout the gray and white matter and generate myelinating cells. OPCs in white matter proliferate more than those in gray matter in response to platelet-derived growth factor AA (PDGF AA), despite similar levels of its alpha receptor (PDGFRα) on their surface. Here we show that the type 1 integral membrane protein neuropilin-1 (Nrp1) is expressed not on OPCs but on amoeboid and activated microglia in white but not gray matter in an age- and activity-dependent manner. Microglia-specific deletion of Nrp1 compromised developmental OPC proliferation in white matter as well as OPC expansion and subsequent myelin repair after acute demyelination. Exogenous Nrp1 increased PDGF AA-induced OPC proliferation and PDGFRα phosphorylation on dissociated OPCs, most prominently in the presence of suboptimum concentrations of PDGF AA. These findings uncover a mechanism of regulating oligodendrocyte lineage cell density that involves trans-activation of PDGFRα on OPCs via Nrp1 expressed by adjacent microglia.
Conflict of interest statement
The authors declare no competing interests.
Figures
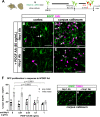
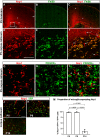
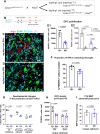
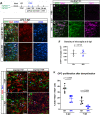
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

