The VH framework region 1 as a target of efficient mutagenesis for generating a variety of affinity-matured scFv mutants
- PMID: 33859250
- PMCID: PMC8050046
- DOI: 10.1038/s41598-021-87501-7
The VH framework region 1 as a target of efficient mutagenesis for generating a variety of affinity-matured scFv mutants
Abstract
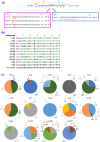
In vitro affinity-maturation potentially generates antibody fragments with enhanced antigen-binding affinities that allow for developing more sensitive diagnostic systems and more effective therapeutic agents. Site-directed mutagenesis targeting "hot regions," i.e., amino acid substitutions therein frequently increase the affinities, is desirable for straightforward discovery of valuable mutants. We here report two "designed" site-directed mutagenesis (A and B) targeted the N-terminal 1-10 positions of the VH framework region 1 that successfully improved an anti-cortisol single-chain Fv fragment (Ka, 3.6 × 108 M-1). Mutagenesis A substituted the amino acids at the position 1-3, 5-7, 9 and 10 with a limited set of substitutions to generate only 1,536 different members, while mutagenesis B inserted 1-6 random residues between the positions 6 and 7. Screening the resulting bacterial libraries as scFv-phage clones with a clonal array profiling system provided 21 genetically unique scFv mutants showing 17-31-fold increased affinity with > 109 M-1 Ka values. Among the mutants selected from the library A and B, scFv mA#18 (with five-residue substitutions) and mB1-3#130 (with a single residue insertion) showed the greatest Ka value, 1.1 × 1010 M-1.
Conflict of interest statement
The authors declare no competing interests.
Figures


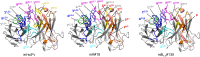

References
-
- An Z, editor. Therapeutic Monoclonal Antibodies, from Bench to Clinic. New York: Wiley; 2009.
-
- Dübel S, editor. Handbook of Therapeutic Antibodies. New York: Wiley-Blackwell; 2010.
-
- Wild D, editor. The Immunoassay Handbook. Amsterdam: Elsevier; 2013.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

