Activation of creER recombinase in the mouse calvaria induces local recombination without effects on distant skeletal segments
- PMID: 33859263
- PMCID: PMC8050205
- DOI: 10.1038/s41598-021-87611-2
Activation of creER recombinase in the mouse calvaria induces local recombination without effects on distant skeletal segments
Abstract
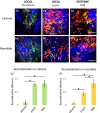
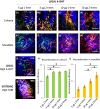
Conditional creER-mediated gene inactivation or gene induction has emerged as a robust tool for studying gene functions in mouse models of tissue development, homeostasis, and regeneration. Here, we present a method to conditionally induce cre recombination in the mouse calvarial bone while avoiding systemic recombination in distal bones. To test our method, we utilized Prx1creER-egfp;td-Tomato mice and delivered 4-hydroxytamoxifen (4-OHT) to the mouse calvaria, subperiosteally. First, we showed that two calvaria subperiosteal injections of 10 µg of 4-OHT (3.3 mg of 4-OHT/kg of body weight) can induce local recombination as efficiently as two intraperitoneal systemic injections of 200 μg of tamoxifen (70 mg of tamoxifen/kg of body weight). Then, we studied the recombination efficiency of various subperiosteal calvaria dosages and found that two subperiosteal injections of 5 µg 4-OHT (1.65 mg of 4-OHT/kg of body weight) uphold the same recombination efficiency observed with higher dosages. Importantly, the result indicated that the low dosage does not induce significant systemic recombination in remote skeletal tissues. With the proposed local low dosage protocol, the recombination efficiency at the injection site (calvarial bone) reached 94%, while the recombination efficiency at the mandible and the digits was as low as the efficiency measured in control animals.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Kühn, R. & Wurst, W. Gene knockout protocols. 2nd ed. In Methods in molecular biology, xvi, 4 p. of plates (Humana Press, 2009).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

