Estradiol analogs attenuate autophagy, cell migration and invasion by direct and selective inhibition of TRPML1, independent of estrogen receptors
- PMID: 33859333
- PMCID: PMC8050276
- DOI: 10.1038/s41598-021-87817-4
Estradiol analogs attenuate autophagy, cell migration and invasion by direct and selective inhibition of TRPML1, independent of estrogen receptors
Abstract
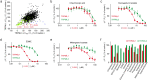
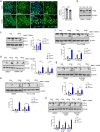
The cation channel TRPML1 is an important regulator of lysosomal function and autophagy. Loss of TRPML1 is associated with neurodegeneration and lysosomal storage disease, while temporary inhibition of this ion channel has been proposed to be beneficial in cancer therapy. Currently available TRPML1 channel inhibitors are not TRPML isoform selective and block at least two of the three human isoforms. We have now identified the first highly potent and isoform-selective TRPML1 antagonist, the steroid 17β-estradiol methyl ether (EDME). Two analogs of EDME, PRU-10 and PRU-12, characterized by their reduced activity at the estrogen receptor, have been identified through systematic chemical modification of the lead structure. EDME and its analogs, besides being promising new small molecule tool compounds for the investigation of TRPML1, selectively affect key features of TRPML1 function: autophagy induction and transcription factor EB (TFEB) translocation. In addition, they act as inhibitors of triple-negative breast cancer cell migration and invasion.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Tsunemi T, Perez-Rosello T, Ishiguro Y, Yoroisaka A, Jeon S, Hamada K, Rammonhan M, Wong YC, Xie Z, Akamatsu W, et al. Increased lysosomal exocytosis induced by lysosomal Ca(2+) channel agonists protects human dopaminergic neurons from alpha-synuclein toxicity. J. Neurosci. 2019;39:5760–5772. doi: 10.1523/JNEUROSCI.3085-18.2019. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

