ZNF410 represses fetal globin by singular control of CHD4
- PMID: 33859416
- PMCID: PMC8180380
- DOI: 10.1038/s41588-021-00843-w
ZNF410 represses fetal globin by singular control of CHD4
Abstract
Known fetal hemoglobin (HbF) silencers have potential on-target liabilities for rational β-hemoglobinopathy therapeutic inhibition. Here, through transcription factor (TF) CRISPR screening, we identify zinc-finger protein (ZNF) 410 as an HbF repressor. ZNF410 does not bind directly to the genes encoding γ-globins, but rather its chromatin occupancy is concentrated solely at CHD4, encoding the NuRD nucleosome remodeler, which is itself required for HbF repression. CHD4 has two ZNF410-bound regulatory elements with 27 combined ZNF410 binding motifs constituting unparalleled genomic clusters. These elements completely account for the effects of ZNF410 on fetal globin repression. Knockout of ZNF410 or its mouse homolog Zfp410 reduces CHD4 levels by 60%, enough to substantially de-repress HbF while eluding cellular or organismal toxicity. These studies suggest a potential target for HbF induction for β-hemoglobin disorders with a wide therapeutic index. More broadly, ZNF410 represents a special class of gene regulator, a conserved TF with singular devotion to regulation of a chromatin subcomplex.
Conflict of interest statement
Competing interests statement
D.E.B. and D.S.V. are co-inventors on a patent related to ZNF410 disruption. The authors declare no other competing interests.
Figures



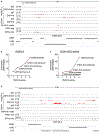




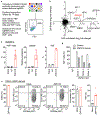

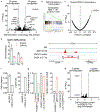


Comment in
-
Targeting ZNF410 as a potential β-hemoglobinopathy therapy.Nat Genet. 2021 May;53(5):589-590. doi: 10.1038/s41588-021-00817-y. Nat Genet. 2021. PMID: 33859418 Free PMC article.
References
-
- Orkin SH & Bauer DE Emerging Genetic Therapy for Sickle Cell Disease. Annu. Rev. Med 70, 257–271 (2019). - PubMed
-
- Piel FB, Steinberg MH & Rees DC Sickle Cell Disease. The New England journal of medicine vol. 377 305 (2017). - PubMed
-
- Weatherall DJ The Evolving Spectrum of the Epidemiology of Thalassemia. Hematol. Oncol. Clin. North Am 32, 165–175 (2018). - PubMed
-
- Sankaran VG et al. Human Fetal Hemoglobin Expression Is Regulated by the Developmental Stage-Specific Repressor BCL11A. Science 322, 1839–1842 (2008). - PubMed
References for Methods
-
- Vinjamur DS & Bauer DE Growing and Genetically Manipulating Human Umbilical Cord Blood-Derived Erythroid Progenitor (HUDEP) Cell Lines. Methods Mol. Biol 1698, 275–284 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

