Estrogen Receptor Beta Influences the Inflammatory p65 Cistrome in Colon Cancer Cells
- PMID: 33859619
- PMCID: PMC8042384
- DOI: 10.3389/fendo.2021.650625
Estrogen Receptor Beta Influences the Inflammatory p65 Cistrome in Colon Cancer Cells
Abstract
Inflammation is a primary component of both initiation and promotion of colorectal cancer (CRC). Cytokines secreted by macrophages, including tumor necrosis factor alpha (TNFα), activates the pro-survival transcription factor complex NFκB. The precise mechanism of NFκB in CRC is not well studied, but we recently reported the genome-wide transcriptional impact of TNFα in two CRC cell lines. Further, estrogen signaling influences inflammation in a complex manner and suppresses CRC development. CRC protective effects of estrogen have been shown to be mediated by estrogen receptor beta (ERβ, ESR2), which also impacts inflammatory signaling of the colon. However, whether ERβ impacts the chromatin interaction (cistrome) of the main NFκB subunit p65 (RELA) is not known. We used p65 chromatin immunoprecipitation followed by sequencing (ChIP-Seq) in two different CRC cell lines, HT29 and SW480, with and without expression of ERβ. We here present the p65 colon cistrome of these two CRC cell lines. We identify that RELA and AP1 motifs are predominant in both cell lines, and additionally describe both common and cell line-specific p65 binding sites and correlate these to transcriptional changes related to inflammation, migration, apoptosis and circadian rhythm. Further, we determine that ERβ opposes a major fraction of p65 chromatin binding in HT29 cells, but enhances p65 binding in SW480 cells, thereby impacting the p65 cistrome differently in the two cell lines. However, the biological functions of the regulated genes appear to have similar roles in both cell lines. To our knowledge, this is the first time the p65 CRC cistrome is compared between different cell lines and the first time an influence by ERβ on the p65 cistrome is investigated. Our work provides a mechanistic foundation for a better understanding of how estrogen influences inflammatory signaling through NFκB in CRC cells.
Keywords: ChIP; ERβ; colon cancer; colorectal cancer (CRC); estrogen receptor; p65.
Copyright © 2021 Indukuri, Hases, Archer and Williams.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

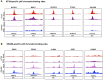



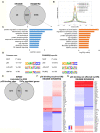

References
-
- Zheng SL, Roddick AJ. Association of Aspirin Use for Primary Prevention With Cardiovascular Events and Bleeding Events: A Systematic Review and Meta-analysisAssociation of Aspirin Use for Primary Prevention of CVD With Cardiovascular Events and BleedingAssociation of Aspirin Use for Primary Prevention of CVD With Cardiovascular Events and Bleeding. JAMA (2019) 321(3):277–87. 10.1001/jama.2018.20578 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

