Characterization and Mechanism of Linearized-Microcystinase Involved in Bacterial Degradation of Microcystins
- PMID: 33859631
- PMCID: PMC8042282
- DOI: 10.3389/fmicb.2021.646084
Characterization and Mechanism of Linearized-Microcystinase Involved in Bacterial Degradation of Microcystins
Abstract
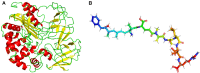
Microcystins (MCs) are extremely hazardous to the ecological environment and public health. How to control and remove MCs is an unsolved problem all over the world. Some microbes and their enzymes are thought to be effective in degrading MCs. Microcystinase can linearize microcystin-leucine-arginine (MC-LR) via a specific locus. However, linearized MC-LR is also very toxic and needs to be removed. How linearized MC-LR was metabolized by linearized-microcystinase, especially how linearized-microcystinase binds to linearized MC-LR, has not been defined. A combination of in vitro experiments and computer simulation was applied to explore the characterization and molecular mechanisms for linearized MC-LR degraded by linearized-microcystinase. The purified linearized-microcystinase was obtained by recombinant Escherichia coli overexpressing. The concentration of linearized MC-LR was detected by high-performance liquid chromatography, and linearized MC-LR degradation products were analyzed by the mass spectrometer. Homology modeling was used to predict the structure of the linearized-microcystinase. Molecular docking techniques on the computer were used to simulate the binding sites of linearized-microcystinase and linearized MC-LR. The purified linearized-microcystinase was obtained successfully. The linearized-microcystinase degraded linearized MC-LR to tetrapeptide efficiently. The second structure of linearized-microcystinase consisted of many alpha-helices, beta-strands, and colis. Linearized-microcystinase interacted the linearized MC-LR with hydrogen bond, hydrophobic interaction, electrostatic forces, and the Van der Waals force. This study firstly reveals the characterization and specific enzymatic mechanism of linearized-microcystinase for catalyzing linearized MC-LR. These findings encourage the application of MC-degrading engineering bacteria and build a great technique for MC-LR biodegradation in environmental engineering.
Keywords: biodegradation; homology modeling; linearized-microcystinase; microcystins; molecular docking.
Copyright © 2021 Wei, Huang, Feng, Massey, Clara, Long, Cao, Luo and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Carmichael W. W., Meriluoto J., Spoof L., Codd G. A. (2018). Handbook of cyanobacterial monitoring and cyanotoxin analysis. Anal. Bioanal. Chem. 410 1405–1406. 10.1007/s00216-017-0804-x - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

