Disparate Interferon Signaling and Shared Aberrant Basaloid Cells in Single-Cell Profiling of Idiopathic Pulmonary Fibrosis and Systemic Sclerosis-Associated Interstitial Lung Disease
- PMID: 33859634
- PMCID: PMC8042271
- DOI: 10.3389/fimmu.2021.595811
Disparate Interferon Signaling and Shared Aberrant Basaloid Cells in Single-Cell Profiling of Idiopathic Pulmonary Fibrosis and Systemic Sclerosis-Associated Interstitial Lung Disease
Abstract
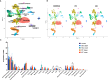
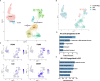
Idiopathic pulmonary fibrosis (IPF) and systemic sclerosis-associated interstitial lung disease (SSc-ILD) differ in the predominant demographics and identified genetic risk alleles of effected patients, however both diseases frequently progress to respiratory failure and death. Contrasting advanced SSc-ILD to IPF provides insight to the role dysregulated immunity may play in pulmonary fibrosis. To analyze cell-type specific transcriptome commonalities and differences between IPF and SSc-ILD, we compared single-cell RNA-sequencing (scRNA-seq) of 21 explanted lung tissue specimens from patients with advanced IPF, SSc-ILD, and organ donor controls. Comparison of IPF and SSc-ILD tissue identified divergent patterns of interferon signaling, with interferon-gamma signaling upregulated in the SPP1hi and FABP4hi macrophages, cytotoxic T cells, and natural kill cells of IPF, while type I interferon signaling and production was upregulated in the corresponding SSc-ILD populations. Plasmacytoid dendritic cells were found in diseased lungs only, and exhibited upregulated cellular stress pathways in SSc-ILD compared to IPF. Alveolar type I cells were dramatically decreased in both IPF and SSc-ILD, with a distinct transcriptome signature separating these cells by disease. KRT5-/KRT17+ aberrant basaloid cells exhibiting markers of cellular senescence and epithelial-mesenchymal transition were identified in SSc-ILD for the first time. In summary, our study utilizes the enriched capabilities of scRNA-seq to identify key divergent cell types and pathways between IPF and SSc-ILD, providing new insights into the shared and distinct mechanisms between idiopathic and autoimmune interstitial lung diseases.
Keywords: idiopathic pulmonary fibrosis; interstitial lung disease (ILD); single-cell RNA-sequencing (scRNA-seq); systemic sclerosis; systemic sclerosis (scleroderma).
Copyright © 2021 Valenzi, Tabib, Papazoglou, Sembrat, Trejo Bittar, Rojas and Lafyatis.
Conflict of interest statement
RL reports grants from Bristol Myers Squibb, Corbus, Formation, Elpidera, Regeneron, Pfizer, and Kiniksa outside the submitted work; personal fees from Bristol Myers Squibb, Formation, Sanofi, Biocon, Boehringer Mannheim, Merck, and Genentech/Roche outside the submitted work. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

